1 c3bk6C_
99.9
42
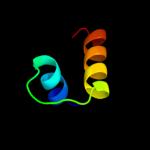
PDB header: membrane proteinChain: C: PDB Molecule: ph stomatin;PDBTitle: crystal structure of a core domain of stomatin from2 pyrococcus horikoshii
2 c2rpbA_
99.8
50
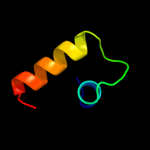
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical membrane protein;PDBTitle: the solution structure of membrane protein
3 d1wina_
99.8
15
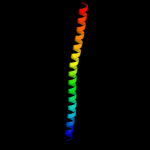
Fold: EF-Ts domain-likeSuperfamily: Band 7/SPFH domainFamily: Band 7/SPFH domain4 c2zv4O_
98.4
16
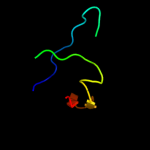
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
5 c3u5gB_
58.4
19
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein s1-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
6 c2xzm4_
43.2
11
PDB header: ribosomeChain: 4: PDB Molecule: 40s ribosomal protein s3a;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
7 c3k5bB_
42.1
21
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase, subunit (vapc-therm);PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
8 c1y4cA_
23.5
16
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
9 d1nkta1
21.3
31
Fold: Pre-protein crosslinking domain of SecASuperfamily: Pre-protein crosslinking domain of SecAFamily: Pre-protein crosslinking domain of SecA10 d1tf5a1
18.9
31
Fold: Pre-protein crosslinking domain of SecASuperfamily: Pre-protein crosslinking domain of SecAFamily: Pre-protein crosslinking domain of SecA11 d2hsga1
12.8
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator12 d1lcda_
11.8
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator13 d2bjca1
10.8
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator14 c2lcvA_
10.8
15
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional repressor cytr;PDBTitle: structure of the cytidine repressor dna-binding domain; an alternate2 calculation
15 c2k88A_
10.5
15
PDB header: hydrolaseChain: A: PDB Molecule: vacuolar proton pump subunit g;PDBTitle: association of subunit d (vma6p) and e (vma4p) with g2 (vma10p) and the nmr solution structure of subunit g (g1-3 59) of the saccharomyces cerevisiae v1vo atpase
16 c2l8nA_
9.6
15
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional repressor cytr;PDBTitle: nmr structure of the cytidine repressor dna binding domain in presence2 of operator half-site dna
17 d1qpza1
9.2
6
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator18 d1efaa1
8.7
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator19 d1l2pa_
8.4
31
Fold: Single transmembrane helixSuperfamily: F1F0 ATP synthase subunit B, membrane domainFamily: F1F0 ATP synthase subunit B, membrane domain20 d2f1la1
8.1
17
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: RimM C-terminal domain-like21 c1wd6B_
not modelled
7.9
38
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein ydhr;PDBTitle: crystal structure of jw1657 from escherichia coli
22 c2kk7A_
not modelled
7.9
23
PDB header: hydrolaseChain: A: PDB Molecule: v-type atp synthase subunit e;PDBTitle: nmr solution structure of the n terminal domain of subunit e2 (e1-52) of a1ao atp synthase from methanocaldococcus3 jannaschii
23 c3lj5H_
not modelled
7.6
15
PDB header: viral proteinChain: H: PDB Molecule: portal protein;PDBTitle: full length bacteriophage p22 portal protein
24 c3sbtB_
not modelled
7.1
8
PDB header: splicingChain: B: PDB Molecule: a1 cistron-splicing factor aar2;PDBTitle: crystal structure of a aar2-prp8 complex
25 c2ptmA_
not modelled
6.9
17
PDB header: transport proteinChain: A: PDB Molecule: hyperpolarization-activated (ih) channel;PDBTitle: structure and rearrangements in the carboxy-terminal region of spih2 channels
26 c1o7fA_
not modelled
6.8
21
PDB header: regulationChain: A: PDB Molecule: camp-dependent rap1 guanine-nucleotide exchangePDBTitle: crystal structure of the regulatory domain of epac2
27 c1wrgA_
not modelled
6.8
22
PDB header: membrane proteinChain: A: PDB Molecule: light-harvesting protein b-880, beta chain;PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
28 c3sokB_
not modelled
6.6
22
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
29 c3h5tA_
not modelled
6.5
6
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of a transcriptional regulator, lacl2 family protein from corynebacterium glutamicum
30 c2jp3A_
not modelled
6.4
28
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
31 c3arcl_
not modelled
6.3
12
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
32 c3arcL_
not modelled
6.2
12
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
33 c1s5lL_
not modelled
6.2
12
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
34 c3kziL_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
35 c2axtl_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
36 c3bz1L_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
37 c3a0hl_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
38 c3a0bl_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
39 c3a0hL_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
40 c3prqL_
not modelled
6.2
12
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
41 c3prrL_
not modelled
6.2
12
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
42 c1s5ll_
not modelled
6.2
12
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
43 d2axtl1
not modelled
6.2
12
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like44 c3bz2L_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
45 c3a0bL_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
46 c2axtL_
not modelled
6.2
12
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
47 c2jo1A_
not modelled
6.1
28
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
48 d1uxda_
not modelled
6.0
3
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator49 d2pila_
not modelled
5.9
33
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin50 d2ix0a1
not modelled
5.9
28
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like51 d2h8pc1
not modelled
5.8
12
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels52 c3f6sI_
not modelled
5.6
13
PDB header: electron transportChain: I: PDB Molecule: flavodoxin;PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
53 d1qd1a2
not modelled
5.6
9
Fold: Ferredoxin-likeSuperfamily: Formiminotransferase domain of formiminotransferase-cyclodeaminase.Family: Formiminotransferase domain of formiminotransferase-cyclodeaminase.54 d2ha9a1
not modelled
5.4
14
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: SP0239-like55 d1oqwa_
not modelled
5.4
17
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin56 c2opkC_
not modelled
5.3
23
PDB header: isomeraseChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
57 d1i9za_
not modelled
5.3
13
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Inositol polyphosphate 5-phosphatase (IPP5)58 c2f1lA_
not modelled
5.2
23
PDB header: unknown functionChain: A: PDB Molecule: 16s rrna processing protein;PDBTitle: crystal structure of a putative 16s ribosomal rna processing protein2 rimm (pa3744) from pseudomonas aeruginosa at 2.46 a resolution
59 d1jeya1
not modelled
5.1
41
Fold: SPOC domain-likeSuperfamily: SPOC domain-likeFamily: Ku70 subunit middle domain60 c1zvvA_
not modelled
5.1
9
PDB header: transcription/dnaChain: A: PDB Molecule: glucose-resistance amylase regulator;PDBTitle: crystal structure of a ccpa-crh-dna complex