| 1 |
|
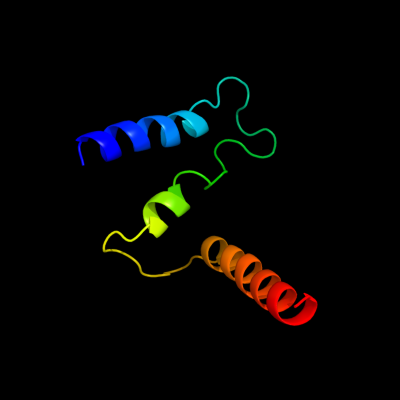
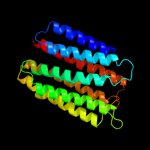
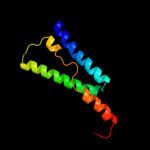
PDB 1s7b chain A
Region: 220 - 290
Aligned: 71
Modelled: 71
Confidence: 97.7%
Identity: 15%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 2 |
|
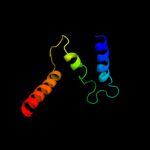
PDB 2i68 chain B
Region: 224 - 287
Aligned: 58
Modelled: 64
Confidence: 89.2%
Identity: 24%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 3 |
|
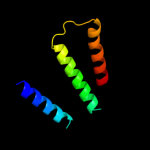
PDB 1xio chain A
Region: 33 - 240
Aligned: 208
Modelled: 208
Confidence: 24.3%
Identity: 5%
PDB header:signaling protein
Chain: A: PDB Molecule:anabaena sensory rhodopsin;
PDBTitle: anabaena sensory rhodopsin
Phyre2
| 4 |
|
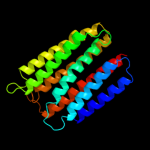
PDB 1xio chain A
Region: 33 - 240
Aligned: 208
Modelled: 208
Confidence: 24.3%
Identity: 5%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 5 |
|
PDB 2jp3 chain A
Region: 272 - 299
Aligned: 28
Modelled: 28
Confidence: 12.5%
Identity: 18%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 6 |
|
PDB 2rdd chain B
Region: 268 - 297
Aligned: 30
Modelled: 30
Confidence: 7.7%
Identity: 7%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 7 |
|
PDB 1w8x chain P
Region: 265 - 299
Aligned: 32
Modelled: 32
Confidence: 7.4%
Identity: 25%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 8 |
|
PDB 2jwa chain A
Region: 269 - 297
Aligned: 29
Modelled: 29
Confidence: 7.4%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 9 |
|
PDB 3aqp chain B
Region: 212 - 299
Aligned: 88
Modelled: 88
Confidence: 6.6%
Identity: 9%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2