| 1 |
|
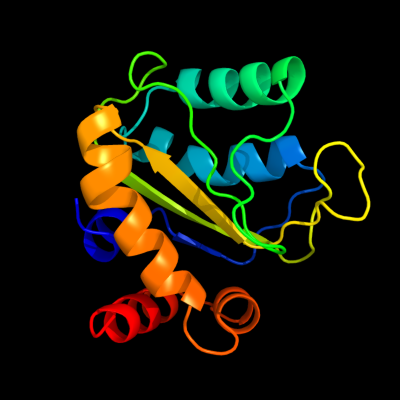
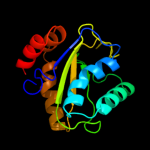
PDB 3bjv chain A
Region: 1 - 153
Aligned: 151
Modelled: 151
Confidence: 100.0%
Identity: 43%
PDB header:transferase
Chain: A: PDB Molecule:rmpa;
PDBTitle: the crystal structure of a putative pts iia(ptxa) from streptococcus2 mutans
Phyre2
| 2 |
|
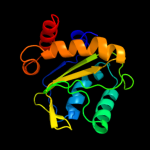
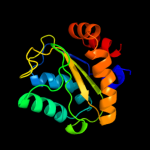
PDB 2oqt chain D
Region: 2 - 153
Aligned: 150
Modelled: 150
Confidence: 100.0%
Identity: 42%
PDB header:transferase
Chain: D: PDB Molecule:hypothetical protein spy0176;
PDBTitle: structural genomics, the crystal structure of a putative2 pts iia domain from streptococcus pyogenes m1 gas
Phyre2
| 3 |
|
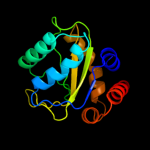
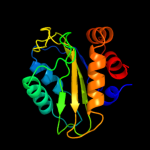
PDB 3oxp chain B
Region: 3 - 150
Aligned: 147
Modelled: 147
Confidence: 100.0%
Identity: 36%
PDB header:transferase
Chain: B: PDB Molecule:phosphotransferase enzyme ii, a component;
PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
Phyre2
| 4 |
|
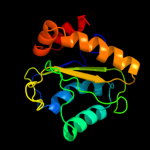
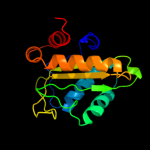
PDB 3oxp chain A
Region: 3 - 150
Aligned: 147
Modelled: 147
Confidence: 100.0%
Identity: 36%
PDB header:transferase
Chain: A: PDB Molecule:phosphotransferase enzyme ii, a component;
PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
Phyre2
| 5 |
|
PDB 2oq3 chain A
Region: 1 - 149
Aligned: 146
Modelled: 149
Confidence: 100.0%
Identity: 36%
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific cryptic phosphotransferase
PDBTitle: solution structure of the mannitol- specific cryptic2 phosphotransferase enzyme iia cmtb from escherichia coli
Phyre2
| 6 |
|
PDB 3urr chain B
Region: 1 - 151
Aligned: 147
Modelled: 151
Confidence: 100.0%
Identity: 18%
PDB header:transferase
Chain: B: PDB Molecule:pts iia-like nitrogen-regulatory protein ptsn;
PDBTitle: structure of pts iia-like nitrogen-regulatory protein ptsn (bth_i0484)2 (ptsn)
Phyre2
| 7 |
|
PDB 1a6j chain A
Region: 1 - 151
Aligned: 148
Modelled: 151
Confidence: 100.0%
Identity: 24%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 8 |
|
PDB 1a3a chain A
Region: 8 - 148
Aligned: 137
Modelled: 141
Confidence: 100.0%
Identity: 31%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 9 |
|
PDB 2a0j chain A
Region: 2 - 147
Aligned: 143
Modelled: 146
Confidence: 100.0%
Identity: 17%
PDB header:transferase
Chain: A: PDB Molecule:pts system, nitrogen regulatory iia protein;
PDBTitle: crystal structure of nitrogen regulatory protein iia-ntr from2 neisseria meningitidis
Phyre2
| 10 |
|
PDB 1xiz chain A
Region: 3 - 151
Aligned: 147
Modelled: 149
Confidence: 100.0%
Identity: 18%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 11 |
|
PDB 1hyn chain P
Region: 4 - 150
Aligned: 143
Modelled: 147
Confidence: 97.6%
Identity: 13%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: Anion transport protein, cytoplasmic domain
Phyre2
| 12 |
|
PDB 1hyn chain Q
Region: 4 - 150
Aligned: 146
Modelled: 147
Confidence: 97.5%
Identity: 15%
PDB header:membrane protein
Chain: Q: PDB Molecule:band 3 anion transport protein;
PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
Phyre2
| 13 |
|
PDB 1k4n chain A
Region: 80 - 110
Aligned: 28
Modelled: 31
Confidence: 11.4%
Identity: 14%
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family: Hypothetical protein YecM (EC4020)
Phyre2
| 14 |
|
PDB 2xvs chain A
Region: 54 - 89
Aligned: 36
Modelled: 36
Confidence: 10.6%
Identity: 25%
PDB header:antitumor protein
Chain: A: PDB Molecule:tetratricopeptide repeat protein 5;
PDBTitle: crystal structure of human ttc5 (strap) c-terminal ob2 domain
Phyre2
| 15 |
|
PDB 1gtd chain A
Region: 11 - 41
Aligned: 31
Modelled: 31
Confidence: 7.8%
Identity: 13%
Fold: PurS-like
Superfamily: PurS-like
Family: PurS subunit of FGAM synthetase
Phyre2
| 16 |
|
PDB 2cs7 chain A domain 1
Region: 61 - 81
Aligned: 21
Modelled: 21
Confidence: 6.7%
Identity: 24%
Fold: IL8-like
Superfamily: PhtA domain-like
Family: PhtA domain-like
Phyre2
| 17 |
|
PDB 1ydh chain A
Region: 100 - 150
Aligned: 51
Modelled: 49
Confidence: 6.3%
Identity: 8%
Fold: MCP/YpsA-like
Superfamily: MCP/YpsA-like
Family: MoCo carrier protein-like
Phyre2
| 18 |
|
PDB 3b0v chain D
Region: 128 - 151
Aligned: 24
Modelled: 24
Confidence: 6.1%
Identity: 13%
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
Phyre2
| 19 |
|
PDB 2hnt chain E
Region: 66 - 93
Aligned: 28
Modelled: 28
Confidence: 5.3%
Identity: 29%
PDB header:serine protease
Chain: E: PDB Molecule:gamma-thrombin;
PDBTitle: crystallographic structure of human gamma-thrombin
Phyre2