1 c2r6gF_
100.0
13
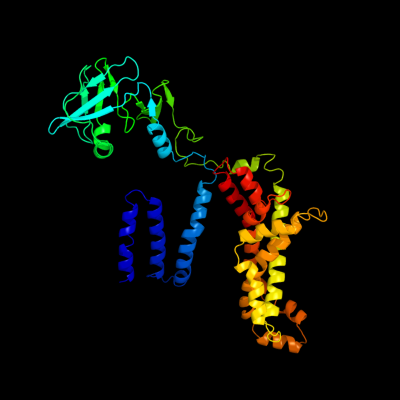
PDB header: hydrolase/transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: the crystal structure of the e. coli maltose transporter
2 c3d31D_
100.0
19
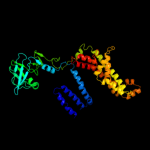
PDB header: transport proteinChain: D: PDB Molecule: sulfate/molybdate abc transporter, permeasePDBTitle: modbc from methanosarcina acetivorans
3 d3d31c1
100.0
19
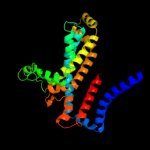
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like4 c3fh6F_
100.0
16
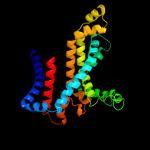
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
5 d2onkc1
100.0
22
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like6 c2onkC_
100.0
22
PDB header: membrane proteinChain: C: PDB Molecule: molybdate/tungstate abc transporter, permeasePDBTitle: abc transporter modbc in complex with its binding protein2 moda
7 d2r6gf2
100.0
16
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like8 d2r6gg1
99.9
19
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like9 d3dhwa1
99.8
16
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like10 c1umqA_
49.9
16
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
11 d1umqa_
49.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like12 d1ntca_
48.9
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like13 d1fipa_
41.1
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like14 c3e7lD_
34.6
13
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
15 d1etob_
34.1
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like16 d1etxa_
30.3
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like17 c2hx6A_
29.5
15
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
18 d1g2ha_
25.7
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like19 c2jwaA_
14.5
24
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
20 c3rkoF_
12.0
12
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
21 c2cw1A_
not modelled
9.6
27
PDB header: de novo proteinChain: A: PDB Molecule: sn4m;PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
22 c3mk7F_
not modelled
8.6
23
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
23 d2diia1
not modelled
8.5
11
Fold: BSD domain-likeSuperfamily: BSD domain-likeFamily: BSD domain24 c1pyuD_
not modelled
8.4
15
PDB header: lyaseChain: D: PDB Molecule: aspartate 1-decarboxylase alfa chain;PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
25 d1cf7a_
not modelled
8.2
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp26 c3s1bA_
not modelled
8.0
14
PDB header: signaling proteinChain: A: PDB Molecule: mini-z;PDBTitle: the development of peptide-based tools for the analysis of2 angiogenesis
27 c1loiA_
not modelled
8.0
33
PDB header: hydrolaseChain: A: PDB Molecule: cyclic 3',5'-amp specific phosphodiesterase rd1;PDBTitle: n-terminal splice region of rat c-amp phosphodiesterase,2 nmr, 7 structures
28 c2diiA_
not modelled
8.0
11
PDB header: transcriptionChain: A: PDB Molecule: tfiih basal transcription factor complex p62PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
29 d1v54g_
not modelled
7.8
9
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIaFamily: Mitochondrial cytochrome c oxidase subunit VIa30 c2d7dB_
not modelled
7.7
13
PDB header: hydrolase/dnaChain: B: PDB Molecule: 40-mer from uvrabc system protein b;PDBTitle: structural insights into the cryptic dna dependent atp-ase2 activity of uvrb
31 d1pf4a2
not modelled
7.5
14
Fold: ABC transporter transmembrane regionSuperfamily: ABC transporter transmembrane regionFamily: ABC transporter transmembrane region32 d1st6a6
not modelled
7.4
11
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin33 c1twcF_
not modelled
7.3
29
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iii 23PDBTitle: rna polymerase ii complexed with gtp
34 d1twff_
not modelled
7.3
29
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB635 d1v54c_
not modelled
7.3
11
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like36 c2pmzW_
not modelled
7.1
14
PDB header: translation, transferaseChain: W: PDB Molecule: dna-directed rna polymerase subunit k;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
37 c2kncA_
not modelled
6.7
19
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
38 d2ns0a1
not modelled
6.6
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like39 c1vc3B_
not modelled
6.5
8
PDB header: lyaseChain: B: PDB Molecule: l-aspartate-alpha-decarboxylase heavy chain;PDBTitle: crystal structure of l-aspartate-alpha-decarboxylase
40 d1qkla_
not modelled
6.5
29
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB641 d2axtj1
not modelled
6.2
30
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like42 d1d5ya2
not modelled
6.2
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator43 c2l0kA_
not modelled
6.2
17
PDB header: transcriptionChain: A: PDB Molecule: stage iii sporulation protein d;PDBTitle: nmr solution structure of a transcription factor spoiiid in complex2 with dna
44 d2oy9a1
not modelled
6.1
9
Fold: BH2638-likeSuperfamily: BH2638-likeFamily: BH2638-like45 c3plxB_
not modelled
6.1
15
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: the crystal structure of aspartate alpha-decarboxylase from2 campylobacter jejuni subsp. jejuni nctc 11168
46 c3h0gF_
not modelled
6.0
29
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iiiPDBTitle: rna polymerase ii from schizosaccharomyces pombe
47 d1v54m_
not modelled
6.0
24
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)Family: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)48 c3lk3T_
not modelled
5.9
30
PDB header: protein bindingChain: T: PDB Molecule: leucine-rich repeat-containing protein 16a;PDBTitle: crystal structure of capz bound to the cpi and csi uncapping2 motifs from carmil
49 c2l2tA_
not modelled
5.9
16
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
50 d1sjpa1
not modelled
5.7
13
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: GroEL chaperone, ATPase domain51 c2oviA_
not modelled
5.6
4
PDB header: ligand binding protein, metal transportChain: A: PDB Molecule: hypothetical protein chux;PDBTitle: structure of the heme binding protein chux
52 d2auwa1
not modelled
5.6
10
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like53 d1h6gb1
not modelled
5.5
4
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin54 c2jz1A_
not modelled
5.5
10
PDB header: transcriptionChain: A: PDB Molecule: protein doublesex;PDBTitle: dsx_long
55 d1ppya_
not modelled
5.5
15
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC56 c2vn2B_
not modelled
5.4
8
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
57 d1q3qa1
not modelled
5.4
15
Fold: GroEL equatorial domain-likeSuperfamily: GroEL equatorial domain-likeFamily: Group II chaperonin (CCT, TRIC), ATPase domain58 c1pt1B_
not modelled
5.4
15
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
59 c2lhuA_
not modelled
5.4
10
PDB header: structural proteinChain: A: PDB Molecule: mybpc3 protein;PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
60 c2ks1B_
not modelled
5.3
17
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
61 d3fapb_
not modelled
5.3
8
Fold: Four-helical up-and-down bundleSuperfamily: FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP)Family: FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP)62 c2kvlA_
not modelled
5.2
9
PDB header: viral proteinChain: A: PDB Molecule: major outer capsid protein vp7;PDBTitle: nmr structure of the c-terminal domain of vp7
63 c3qkbB_
not modelled
5.2
5
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function which belongs to2 pfam duf74 family (pepe_0654) from pediococcus pentosaceus atcc 257453 at 2.73 a resolution
64 c2l37A_
not modelled
5.2
18
PDB header: hydrolaseChain: A: PDB Molecule: ribosome-inactivating protein luffin p1;PDBTitle: 3d solution structure of arginine/glutamate-rich polypeptide luffin p12 from the seeds of sponge gourd (luffa cylindrical)
65 c1ql1A_
not modelled
5.1
12
PDB header: virusChain: A: PDB Molecule: pf1 bacteriophage coat protein b;PDBTitle: inovirus (filamentous bacteriophage) strain pf1 major coat2 protein assembly
66 c1uheA_
not modelled
5.1
15
PDB header: lyaseChain: A: PDB Molecule: aspartate 1-decarboxylase alpha chain;PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
67 c2rddB_
not modelled
5.0
17
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
68 c2fomA_
not modelled
5.0
17
PDB header: viral protein/proteaseChain: A: PDB Molecule: polyprotein;PDBTitle: dengue virus ns2b/ns3 protease
69 d1ku7a_
not modelled
5.0
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain70 c2e19A_
not modelled
5.0
9
PDB header: transcriptionChain: A: PDB Molecule: transcription factor 8;PDBTitle: solution structure of the homeobox domain from human nil-2-2 a zinc finger protein, transcription factor 8