| 1 |
|
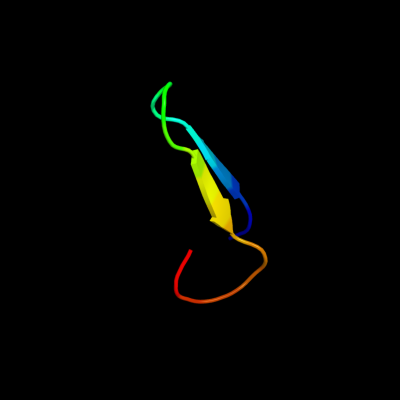
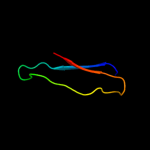
PDB 2knq chain A
Region: 25 - 44
Aligned: 20
Modelled: 20
Confidence: 79.5%
Identity: 40%
PDB header:protein transport
Chain: A: PDB Molecule:general secretion pathway protein h;
PDBTitle: solution structure of e.coli gsph
Phyre2
| 2 |
|
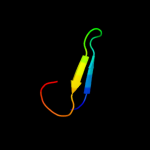
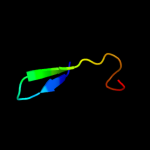
PDB 1qb3 chain B
Region: 29 - 77
Aligned: 45
Modelled: 49
Confidence: 20.8%
Identity: 20%
PDB header:cell cycle
Chain: B: PDB Molecule:cyclin-dependent kinases regulatory subunit;
PDBTitle: crystal structure of the cell cycle regulatory protein cks1
Phyre2
| 3 |
|
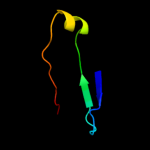
PDB 2vzs chain A domain 2
Region: 35 - 49
Aligned: 15
Modelled: 15
Confidence: 17.3%
Identity: 20%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: beta-Galactosidase/glucuronidase domain
Family: beta-Galactosidase/glucuronidase domain
Phyre2
| 4 |
|
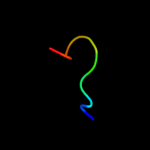
PDB 1drs chain A
Region: 70 - 81
Aligned: 12
Modelled: 12
Confidence: 16.2%
Identity: 50%
Fold: Snake toxin-like
Superfamily: Snake toxin-like
Family: Dendroaspin
Phyre2
| 5 |
|
PDB 1qb3 chain A
Region: 29 - 77
Aligned: 45
Modelled: 49
Confidence: 15.0%
Identity: 20%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 6 |
|
PDB 3pfn chain B
Region: 25 - 53
Aligned: 29
Modelled: 29
Confidence: 11.9%
Identity: 24%
PDB header:transferase
Chain: B: PDB Molecule:nad kinase;
PDBTitle: crystal structure of human nad kinase
Phyre2
| 7 |
|
PDB 1nm8 chain A domain 2
Region: 52 - 83
Aligned: 32
Modelled: 32
Confidence: 7.3%
Identity: 9%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 8 |
|
PDB 2h4t chain B
Region: 53 - 83
Aligned: 31
Modelled: 31
Confidence: 6.6%
Identity: 13%
PDB header:transferase
Chain: B: PDB Molecule:carnitine o-palmitoyltransferase ii,
PDBTitle: crystal structure of rat carnitine palmitoyltransferase ii
Phyre2
| 9 |
|
PDB 3afo chain B
Region: 25 - 50
Aligned: 26
Modelled: 26
Confidence: 6.6%
Identity: 31%
PDB header:transferase
Chain: B: PDB Molecule:nadh kinase pos5;
PDBTitle: crystal structure of yeast nadh kinase complexed with nadh
Phyre2
| 10 |
|
PDB 1u1z chain A
Region: 20 - 29
Aligned: 10
Modelled: 10
Confidence: 6.3%
Identity: 70%
Fold: Thioesterase/thiol ester dehydrase-isomerase
Superfamily: Thioesterase/thiol ester dehydrase-isomerase
Family: FabZ-like
Phyre2
| 11 |
|
PDB 3da7 chain G
Region: 35 - 52
Aligned: 18
Modelled: 18
Confidence: 5.7%
Identity: 39%
PDB header:protein binding
Chain: G: PDB Molecule:barnase circular permutant;
PDBTitle: a conformationally strained, circular permutant of barnase
Phyre2
| 12 |
|
PDB 3bjr chain A
Region: 6 - 40
Aligned: 28
Modelled: 35
Confidence: 5.7%
Identity: 36%
PDB header:hydrolase
Chain: A: PDB Molecule:putative carboxylesterase;
PDBTitle: crystal structure of a putative carboxylesterase (lp_1002) from2 lactobacillus plantarum wcfs1 at 2.09 a resolution
Phyre2
| 13 |
|
PDB 1t1u chain A domain 2
Region: 52 - 83
Aligned: 32
Modelled: 32
Confidence: 5.5%
Identity: 9%
Fold: CoA-dependent acyltransferases
Superfamily: CoA-dependent acyltransferases
Family: Choline/Carnitine O-acyltransferase
Phyre2
| 14 |
|
PDB 2glv chain A
Region: 20 - 29
Aligned: 10
Modelled: 10
Confidence: 5.2%
Identity: 50%
PDB header:lyase
Chain: A: PDB Molecule:(3r)-hydroxymyristoyl-acyl carrier protein
PDBTitle: crystal structure of (3r)-hydroxyacyl-acyl carrier protein2 dehydratase(fabz) mutant(y100a) from helicobacter pylori
Phyre2