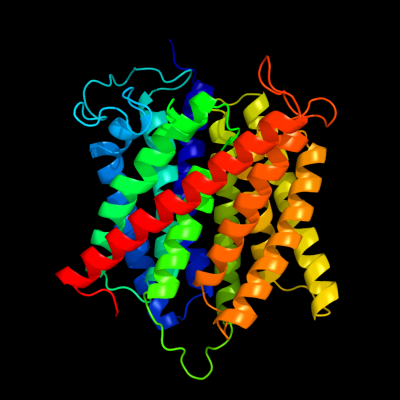
| 1 | c2b2hA_
|
|
 |
100.0 |
39 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
|
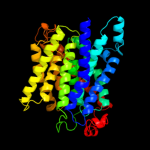
| 2 | d1u7ga_
|
|
 |
100.0 |
96 |
Fold:Ammonium transporter
Superfamily:Ammonium transporter
Family:Ammonium transporter |
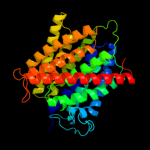
| 3 | c3c1iA_
|
|
 |
100.0 |
99 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonia channel;
PDBTitle: substrate binding, deprotonation and selectivity at the2 periplasmic entrance of the e. coli ammonia channel amtb
|
| 4 | c3hd6A_
|
|
 |
100.0 |
21 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
|
| 5 | c3b9yA_
|
|
 |
100.0 |
23 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
|
| 6 | c3k3gA_
|
|
 |
30.4 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:urea transporter;
PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
|
| 7 | c3qnqD_
|
|
 |
13.1 |
14 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
| 8 | d1u1ha2
|
|
 |
13.0 |
47 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
| 9 | c2kncA_
|
|
 |
12.1 |
25 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 10 | c2nq5A_
|
|
 |
11.8 |
53 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: crystal structure of methyltransferase from streptococcus2 mutans
|
| 11 | d1u1ha1
|
|
 |
11.4 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
| 12 | c3l7sA_
|
|
 |
10.8 |
53 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--homocysteine
PDBTitle: crystal structure of mete coordinated with zinc from streptococcus2 mutans
|
| 13 | c1t7lA_
|
|
 |
10.7 |
67 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: crystal structure of cobalamin-independent methionine2 synthase from t. maritima
|
| 14 | c1u22A_
|
|
 |
10.3 |
47 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: a. thaliana cobalamine independent methionine synthase
|
| 15 | c3lr4A_
|
|
 |
8.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: periplasmic domain of the riss sensor protein from burkholderia2 pseudomallei, barium phased at low ph
|
| 16 | d1z9ha1
|
|
 |
8.6 |
19 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 17 | d1vk5a_
|
|
 |
8.5 |
21 |
Fold:Hypothetical protein At3g22680
Superfamily:Hypothetical protein At3g22680
Family:Hypothetical protein At3g22680 |
| 18 | c1vk5A_
|
|
 |
8.5 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:expressed protein;
PDBTitle: x-ray structure of gene product from arabidopsis thaliana at3g22680
|
| 19 | c3da4B_
|
|
 |
8.4 |
56 |
PDB header:antibiotic
Chain: B: PDB Molecule:colicin-m;
PDBTitle: crystal structure of colicin m, a novel phosphatase2 specifically imported by escherichia coli
|
| 20 | c2dp3A_
|
|
 |
6.3 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
|
| 21 | d1fftb2 |
|
not modelled |
6.2 |
9 |
Fold:Transmembrane helix hairpin
Superfamily:Cytochrome c oxidase subunit II-like, transmembrane region
Family:Cytochrome c oxidase subunit II-like, transmembrane region |
| 22 | d1ttja_ |
|
not modelled |
6.0 |
40 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 23 | d1ezfa_ |
|
not modelled |
6.0 |
28 |
Fold:Terpenoid synthases
Superfamily:Terpenoid synthases
Family:Squalene synthase |
| 24 | c1ru7B_ |
|
not modelled |
5.6 |
28 |
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin;
PDBTitle: 1934 human h1 hemagglutinin
|
| 25 | d1kv5a_ |
|
not modelled |
5.6 |
40 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 26 | c2l9uA_ |
|
not modelled |
5.2 |
33 |
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
|
| 27 | c1mqlB_ |
|
not modelled |
5.1 |
28 |
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin ha2 chain;
PDBTitle: bha of ukr/63
|