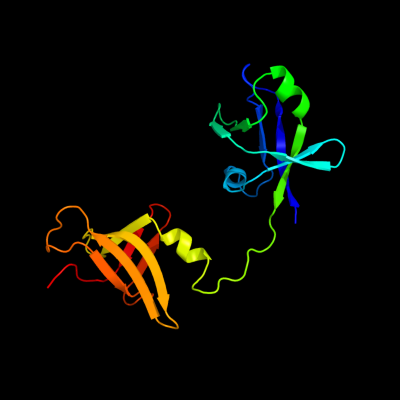
| 1 | c3h9nA_
|
|
 |
100.0 |
66 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:ribosome maturation factor rimm;
PDBTitle: crystal structure of the ribosome maturation factor rimm2 (hi0203) from h.influenzae. northeast structural genomics3 consortium target ir66.
|
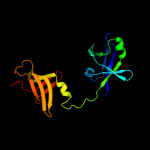
| 2 | c2f1lA_
|
|
 |
100.0 |
38 |
PDB header:unknown function
Chain: A: PDB Molecule:16s rrna processing protein;
PDBTitle: crystal structure of a putative 16s ribosomal rna processing protein2 rimm (pa3744) from pseudomonas aeruginosa at 2.46 a resolution
|
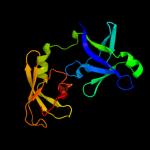
| 3 | c2qggA_
|
|
 |
100.0 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:16s rrna-processing protein rimm;
PDBTitle: x-ray structure of the protein q6f7i0 from acinetobacter2 calcoaceticus amms 248. northeast structural genomics3 consortium target asr73.
|
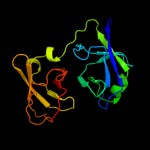
| 4 | c2dyiA_
|
|
 |
100.0 |
30 |
PDB header:ribosome
Chain: A: PDB Molecule:probable 16s rrna-processing protein rimm;
PDBTitle: crystal structure of 16s ribosomal rna processing protein rimm from2 thermus thermophilus hb8
|
| 5 | d2f1la2
|
|
 |
100.0 |
39 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:RimM N-terminal domain-like |
| 6 | c2dogA_
|
|
 |
99.9 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable 16s rrna-processing protein rimm;
PDBTitle: solution structure of the n-terminal domain of rimm from thermus2 thermophilus hb8
|
| 7 | d2f1la1
|
|
 |
99.9 |
39 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:RimM C-terminal domain-like |
| 8 | c3htrB_
|
|
 |
97.9 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized prc-barrel domain protein;
PDBTitle: crystal structure of prc-barrel domain protein from2 rhodopseudomonas palustris
|
| 9 | d1eysh1
|
|
 |
97.4 |
27 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
| 10 | c1eysH_
|
|
 |
97.4 |
27 |
PDB header:electron transport
Chain: H: PDB Molecule:photosynthetic reaction center;
PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
|
| 11 | d1rzhh1
|
|
 |
96.9 |
27 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
| 12 | c1k6nH_
|
|
 |
96.8 |
27 |
PDB header:photosynthesis
Chain: H: PDB Molecule:photosynthetic reaction center h subunit;
PDBTitle: e(l212)a,d(l213)a double mutant structure of photosynthetic reaction2 center from rhodobacter sphaeroides
|
| 13 | d2i5nh1
|
|
 |
96.8 |
24 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
| 14 | c2i5nH_
|
|
 |
96.7 |
24 |
PDB header:photosynthesis
Chain: H: PDB Molecule:reaction center protein h chain;
PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
|
| 15 | d1pm3a_
|
|
 |
94.4 |
22 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:MTH1895 |
| 16 | d1sqra_
|
|
 |
81.4 |
36 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Ribosomal protein L35ae |
| 17 | c4a1dH_
|
|
 |
80.5 |
10 |
PDB header:ribosome
Chain: H: PDB Molecule:rpl35a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
|
| 18 | c3izcj_
|
|
 |
72.0 |
23 |
PDB header:ribosome
Chain: J: PDB Molecule:60s ribosomal protein rpl12 (l11p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 19 | c3iz5j_
|
|
 |
70.9 |
11 |
PDB header:ribosome
Chain: J: PDB Molecule:60s ribosomal protein l12 (l11p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 20 | c2hvyB_
|
|
 |
68.6 |
18 |
PDB header:isomerase/biosynthetic protein/rna
Chain: B: PDB Molecule:small nucleolar rnp similar to gar1;
PDBTitle: crystal structure of an h/aca box rnp from pyrococcus furiosus
|
| 21 | d2ey4c1 |
|
not modelled |
63.4 |
18 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Gar1-like SnoRNP |
| 22 | c3uaiC_ |
|
not modelled |
60.5 |
13 |
PDB header:isomerase/chaperone
Chain: C: PDB Molecule:h/aca ribonucleoprotein complex subunit 1;
PDBTitle: structure of the shq1-cbf5-nop10-gar1 complex from saccharomyces2 cerevisiae
|
| 23 | c2ro0A_ |
|
not modelled |
53.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:histone acetyltransferase esa1;
PDBTitle: solution structure of the knotted tudor domain of the yeast2 histone acetyltransferase, esa1
|
| 24 | c2kk4A_ |
|
not modelled |
40.2 |
34 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein af_2094;
PDBTitle: solution nmr structure of protein af2094 from archaeoglobus2 fulgidus. northeast structural genomics consotium (nesg)3 target gt2
|
| 25 | c2eqnA_ |
|
not modelled |
22.4 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:hypothetical protein loc92345;
PDBTitle: solution structure of the naf1 domain of hypothetical2 protein bc008207 [homo sapiens]
|
| 26 | d1wb1a2 |
|
not modelled |
15.2 |
36 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Elongation factors |
| 27 | d1fxkc_ |
|
not modelled |
13.8 |
27 |
Fold:Long alpha-hairpin
Superfamily:Prefoldin
Family:Prefoldin |
| 28 | d1qhka_ |
|
not modelled |
11.7 |
13 |
Fold:MbtH/L9 domain-like
Superfamily:L9 N-domain-like
Family:N-terminal domain of RNase HI |
| 29 | c3h9wA_ |
|
not modelled |
11.6 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:diguanylate cyclase with pas/pac sensor;
PDBTitle: crystal structure of the n-terminal domain of diguanylate cyclase with2 pas/pac sensor (maqu_2914) from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr66c
|
| 30 | d2b78a1 |
|
not modelled |
10.4 |
21 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Hypothetical RNA methyltransferase domain (HRMD) |
| 31 | c2rnzA_ |
|
not modelled |
9.2 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:histone acetyltransferase esa1;
PDBTitle: solution structure of the presumed chromodomain of the2 yeast histone acetyltransferase, esa1
|
| 32 | c2pdtD_ |
|
not modelled |
9.2 |
15 |
PDB header:circadian clock protein
Chain: D: PDB Molecule:vivid pas protein vvd;
PDBTitle: 2.3 angstrom structure of phosphodiesterase treated vivid
|
| 33 | c2v3mF_ |
|
not modelled |
9.0 |
15 |
PDB header:ribosomal protein
Chain: F: PDB Molecule:naf1;
PDBTitle: structure of the gar1 domain of naf1
|
| 34 | c3ml6D_ |
|
not modelled |
8.8 |
18 |
PDB header:protein transport
Chain: D: PDB Molecule:chimeric complex between protein dishevlled2 homolog dvl-2
PDBTitle: a complex between dishevlled2 and clathrin adaptor ap-2
|
| 35 | d1bywa_ |
|
not modelled |
7.3 |
10 |
Fold:Profilin-like
Superfamily:PYP-like sensor domain (PAS domain)
Family:Flavin-binding PAS domain |
| 36 | c2l4rA_ |
|
not modelled |
7.2 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h member 2;
PDBTitle: nmr solution structure of the n-terminal pas domain of herg
|
| 37 | c3ewkA_ |
|
not modelled |
7.0 |
16 |
PDB header:flavoprotein
Chain: A: PDB Molecule:sensor protein;
PDBTitle: structure of the redox sensor domain of methylococcus capsulatus2 (bath) mmos
|
| 38 | c2gj3A_ |
|
not modelled |
6.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:nitrogen fixation regulatory protein;
PDBTitle: crystal structure of the fad-containing pas domain of the2 protein nifl from azotobacter vinelandii.
|
| 39 | c3mxqC_ |
|
not modelled |
6.1 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:sensor protein;
PDBTitle: crystal structure of sensory box sensor histidine kinase from vibrio2 cholerae
|
| 40 | d1n9la_ |
|
not modelled |
5.8 |
21 |
Fold:Profilin-like
Superfamily:PYP-like sensor domain (PAS domain)
Family:Flavin-binding PAS domain |
| 41 | c3kxrA_ |
|
not modelled |
5.7 |
27 |
PDB header:transport protein
Chain: A: PDB Molecule:magnesium transporter, putative;
PDBTitle: structure of the cystathionine beta-synthase pair domain of the2 putative mg2+ transporter so5017 from shewanella oneidensis mr-1.
|
| 42 | d2f5ka1 |
|
not modelled |
5.6 |
4 |
Fold:SH3-like barrel
Superfamily:Chromo domain-like
Family:Chromo barrel domain |
| 43 | c3gbyA_ |
|
not modelled |
5.6 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ct1051;
PDBTitle: crystal structure of a protein with unknown function ct10512 from chlorobium tepidum
|
| 44 | c2w7aA_ |
|
not modelled |
5.6 |
14 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:line-1 orf1p;
PDBTitle: structure of the human line-1 orf1p central domain
|
| 45 | c3luqC_ |
|
not modelled |
5.4 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:sensor protein;
PDBTitle: the crystal structure of a pas domain from a sensory box2 histidine kinase regulator from geobacter sulfurreducens to3 2.5a
|
| 46 | c2zdiC_ |
|
not modelled |
5.4 |
27 |
PDB header:chaperone
Chain: C: PDB Molecule:prefoldin subunit alpha;
PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
|
| 47 | c3l31B_ |
|
not modelled |
5.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable manganase-dependent inorganic
PDBTitle: crystal structure of the cbs and drtgg domains of the2 regulatory region of clostridium perfringens3 pyrophosphatase complexed with the inhibitor, amp
|
| 48 | c3oi8B_ |
|
not modelled |
5.1 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of functionally unknown conserved protein domain2 from neisseria meningitidis mc58
|