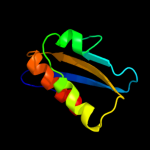
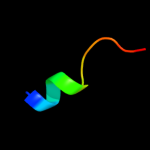
1 d1ufwa_
56.6
23
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD2 d2dy1a3
51.1
29
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components3 c2dnrA_
41.8
19
PDB header: rna binding proteinChain: A: PDB Molecule: synaptojanin-1;PDBTitle: solution structure of rna binding domain in synaptojanin 1
4 d2bv3a3
37.4
21
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components5 c1vbkA_
32.3
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ph1313;PDBTitle: crystal structure of ph1313 from pyrococcus horikoshii ot3
6 d1mg7a1
25.6
12
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Early switch protein XOL-1, N-terminal domain7 d1vbka2
24.1
14
Fold: THUMP domainSuperfamily: THUMP domain-likeFamily: THUMP domain8 c2nscA_
22.1
19
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: structures of and interactions between domains of trigger factor from2 themotoga maritima
9 d1am7a_
21.1
37
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Lambda lysozyme10 c1mg7B_
20.0
13
PDB header: gene regulationChain: B: PDB Molecule: early switch protein xol-1 2.2k splice form;PDBTitle: crystal structure of xol-1
11 c3jvnA_
15.6
27
PDB header: transferaseChain: A: PDB Molecule: acetyltransferase;PDBTitle: crystal structure of the acetyltransferase vf_1542 from vibrio2 fischeri, northeast structural genomics consortium target vfr136
12 d1uwda_
15.4
10
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like13 d2cu6a1
14.0
14
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like14 c3gtyX_
12.8
19
PDB header: chaperone/ribosomal proteinChain: X: PDB Molecule: trigger factor;PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
15 c3ffvA_
12.7
18
PDB header: protein bindingChain: A: PDB Molecule: protein syd;PDBTitle: crystal structure analysis of syd
16 d2nn6h2
12.7
25
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like17 d1ttea1
10.5
31
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain18 d1wina_
10.2
17
Fold: EF-Ts domain-likeSuperfamily: Band 7/SPFH domainFamily: Band 7/SPFH domain19 c2zkqi_
9.6
15
PDB header: ribosomal protein/rnaChain: I: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
20 d1regx_
9.2
17
Fold: Ferredoxin-likeSuperfamily: Translational regulator protein regAFamily: Translational regulator protein regA21 d1z7ma1
not modelled
9.1
9
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain22 d1yd9a1
not modelled
8.8
15
Fold: Macro domain-likeSuperfamily: Macro domain-likeFamily: Macro domain23 c1fwlD_
not modelled
8.7
13
PDB header: transferaseChain: D: PDB Molecule: homoserine kinase;PDBTitle: crystal structure of homoserine kinase
24 d1puga_
not modelled
8.4
18
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like25 d1dwka2
not modelled
8.3
7
Fold: Cyanase C-terminal domainSuperfamily: Cyanase C-terminal domainFamily: Cyanase C-terminal domain26 d1w1oa1
not modelled
8.1
16
Fold: Ferredoxin-likeSuperfamily: FAD-linked oxidases, C-terminal domainFamily: Cytokinin dehydrogenase 127 c3m0zD_
not modelled
7.6
13
PDB header: lyaseChain: D: PDB Molecule: putative aldolase;PDBTitle: crystal structure of putative aldolase from klebsiella2 pneumoniae.
28 c2kq1A_
not modelled
7.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh0266 protein;PDBTitle: solution structure of protein bh0266 from bacillus2 halodurans. northeast structural genomics consortium target3 bhr97a
29 c1zr5B_
not modelled
7.2
14
PDB header: gene regulationChain: B: PDB Molecule: h2afy protein;PDBTitle: crystal structure of the macro-domain of human core histone variant2 macroh2a1.2
30 d1t11a2
not modelled
6.9
32
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain31 c1ybxA_
not modelled
6.6
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
32 c3lnoA_
not modelled
6.5
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
33 d1p97a_
not modelled
6.5
17
Fold: Profilin-likeSuperfamily: PYP-like sensor domain (PAS domain)Family: Hypoxia-inducible factor Hif2a, C-terminal domain34 c2jo7A_
not modelled
6.5
63
PDB header: surface active proteinChain: A: PDB Molecule: glycosylphosphatidylinositol-anchored merozoitePDBTitle: solution structure of the adhesion protein bd37 from2 babesia divergens
35 d1p42a1
not modelled
6.4
30
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC36 c1usvB_
not modelled
6.3
15
PDB header: chaperoneChain: B: PDB Molecule: aha1;PDBTitle: the structure of the complex between aha1 and hsp90
37 c3ew5B_
not modelled
6.2
11
PDB header: rna binding proteinChain: B: PDB Molecule: macro domain of non-structural protein 3;PDBTitle: structure of the tetragonal crystal form of x (adrp) domain2 from fcov
38 d1o75a3
not modelled
6.1
29
Fold: Tp47 lipoprotein, N-terminal domainSuperfamily: Tp47 lipoprotein, N-terminal domainFamily: Tp47 lipoprotein, N-terminal domain39 c3h0gO_
not modelled
6.0
23
PDB header: transcriptionChain: O: PDB Molecule: dna-directed rna polymerase ii subunit rpb3;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
40 d2q3la1
not modelled
5.9
8
Fold: SpoIIaa-likeSuperfamily: SpoIIaa-likeFamily: Sfri0576-like41 d1y14b2
not modelled
5.9
18
Fold: Dodecin subunit-likeSuperfamily: N-terminal, heterodimerisation domain of RBP7 (RpoE)Family: N-terminal, heterodimerisation domain of RBP7 (RpoE)42 c3q71A_
not modelled
5.8
14
PDB header: transferaseChain: A: PDB Molecule: poly [adp-ribose] polymerase 14;PDBTitle: human parp14 (artd8) - macro domain 2 in complex with adenosine-5-2 diphosphoribose
43 c2rpbA_
not modelled
5.7
11
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical membrane protein;PDBTitle: the solution structure of membrane protein
44 c1ta9A_
not modelled
5.6
11
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol dehydrogenase;PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
45 d1v54g_
not modelled
5.5
25
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIaFamily: Mitochondrial cytochrome c oxidase subunit VIa46 d1u0ua2
not modelled
5.4
11
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like47 d1ee0a2
not modelled
5.4
11
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like48 d1usub_
not modelled
5.4
15
Fold: Aha1/BPI domain-likeSuperfamily: Activator of Hsp90 ATPase, Aha1Family: Activator of Hsp90 ATPase, Aha149 c3imkA_
not modelled
5.3
40
PDB header: metal binding proteinChain: A: PDB Molecule: putative molybdenum carrier protein;PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
50 d2ba0a2
not modelled
5.3
17
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: ECR1 N-terminal domain-like51 d1mxaa3
not modelled
5.2
40
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase52 d1wuda1
not modelled
5.1
13
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases