| 1 |
|
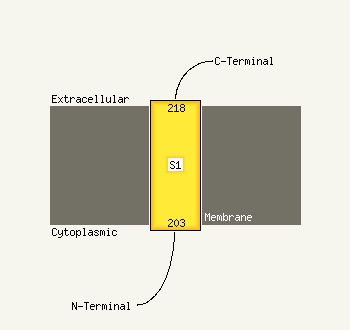
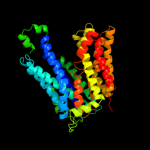
PDB 1pw4 chain A
Region: 3 - 438
Aligned: 417
Modelled: 436
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
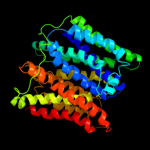
PDB 1pv7 chain A
Region: 20 - 438
Aligned: 400
Modelled: 419
Confidence: 100.0%
Identity: 12%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
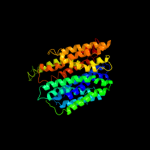
PDB 3o7p chain A
Region: 22 - 435
Aligned: 394
Modelled: 414
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
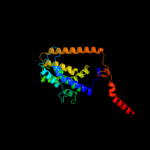
PDB 2gfp chain A
Region: 23 - 426
Aligned: 370
Modelled: 397
Confidence: 100.0%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 27 - 432
Aligned: 397
Modelled: 406
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 11 - 253
Aligned: 224
Modelled: 238
Confidence: 39.1%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 2g9p chain A
Region: 83 - 96
Aligned: 14
Modelled: 14
Confidence: 26.4%
Identity: 43%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 8 |
|
PDB 3hd6 chain A
Region: 7 - 242
Aligned: 220
Modelled: 236
Confidence: 11.5%
Identity: 11%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 9 |
|
PDB 2inp chain D
Region: 84 - 95
Aligned: 12
Modelled: 12
Confidence: 9.9%
Identity: 42%
PDB header:oxidoreductase
Chain: D: PDB Molecule:phenol hydroxylase component phl;
PDBTitle: structure of the phenol hydroxylase-regulatory protein2 complex
Phyre2