| 1 |
|
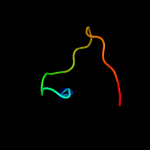
PDB 1avw chain B
Region: 49 - 68
Aligned: 20
Modelled: 20
Confidence: 37.6%
Identity: 40%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Kunitz (STI) inhibitors
Phyre2
| 2 |
|
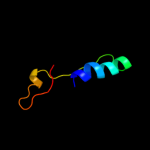
PDB 1eyl chain A
Region: 49 - 68
Aligned: 20
Modelled: 20
Confidence: 34.2%
Identity: 20%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Kunitz (STI) inhibitors
Phyre2
| 3 |
|
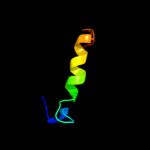
PDB 3bx1 chain C domain 1
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 32.4%
Identity: 28%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Kunitz (STI) inhibitors
Phyre2
| 4 |
|
PDB 3s8j chain B
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 31.6%
Identity: 17%
PDB header:hydrolase inhibitor
Chain: B: PDB Molecule:latex serine proteinase inhibitor;
PDBTitle: crystal structure of a papaya latex serine protease inhibitor (ppi) at2 2.6a resolution
Phyre2
| 5 |
|
PDB 2qn4 chain B
Region: 47 - 68
Aligned: 22
Modelled: 22
Confidence: 27.4%
Identity: 23%
PDB header:hydrolase inhibitor
Chain: B: PDB Molecule:alpha-amylase/subtilisin inhibitor;
PDBTitle: structure and function study of rice bifunctional alpha-2 amylase/subtilisin inhibitor from oryza sativa
Phyre2
| 6 |
|
PDB 2dre chain A
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 27.3%
Identity: 28%
PDB header:plant protein
Chain: A: PDB Molecule:water-soluble chlorophyll protein;
PDBTitle: crystal structure of water-soluble chlorophyll protein from2 lepidium virginicum at 2.00 angstrom resolution
Phyre2
| 7 |
|
PDB 1tie chain A
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 26.6%
Identity: 17%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Kunitz (STI) inhibitors
Phyre2
| 8 |
|
PDB 1wba chain A
Region: 49 - 68
Aligned: 20
Modelled: 20
Confidence: 25.7%
Identity: 30%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Kunitz (STI) inhibitors
Phyre2
| 9 |
|
PDB 1r8n chain A
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 24.9%
Identity: 28%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Kunitz (STI) inhibitors
Phyre2
| 10 |
|
PDB 3iir chain A
Region: 49 - 68
Aligned: 20
Modelled: 20
Confidence: 23.9%
Identity: 15%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:trypsin inhibitor;
PDBTitle: crystal structure of miraculin like protein from seeds of murraya2 koenigii
Phyre2
| 11 |
|
PDB 1r8o chain A
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 14.1%
Identity: 22%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:kunitz trypsin inhibitor;
PDBTitle: crystal structure of an unusual kunitz-type trypsin inhibitor from2 copaifera langsdorffii seeds
Phyre2
| 12 |
|
PDB 2go2 chain A
Region: 51 - 68
Aligned: 18
Modelled: 18
Confidence: 13.5%
Identity: 22%
PDB header:protein binding
Chain: A: PDB Molecule:kunitz-type serine protease inhibitor bbki;
PDBTitle: crystal structure of bbki, a kunitz-type kallikrein inhibitor
Phyre2
| 13 |
|
PDB 1sdo chain A
Region: 24 - 66
Aligned: 42
Modelled: 43
Confidence: 10.6%
Identity: 31%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Restriction endonuclease BstyI
Phyre2
| 14 |
|
PDB 3rnv chain A
Region: 15 - 43
Aligned: 24
Modelled: 29
Confidence: 8.8%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:helper component proteinase;
PDBTitle: structure of the autocatalytic cysteine protease domain of potyvirus2 helper-component proteinase
Phyre2
| 15 |
|
PDB 1agg chain A
Region: 17 - 22
Aligned: 6
Modelled: 6
Confidence: 7.0%
Identity: 100%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: omega toxin-like
Family: Spider toxins
Phyre2
| 16 |
|
PDB 1e88 chain A domain 3
Region: 61 - 66
Aligned: 6
Modelled: 6
Confidence: 5.7%
Identity: 67%
Fold: FnI-like domain
Superfamily: FnI-like domain
Family: Fibronectin type I module
Phyre2