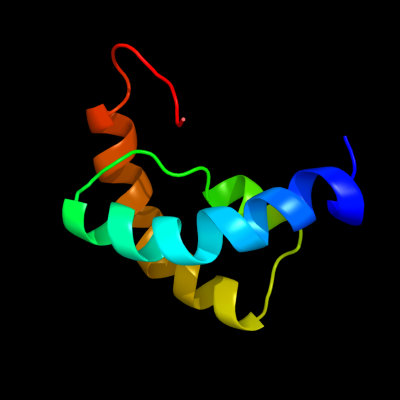
1 d1rrza_
100.0
100
Fold: Spectrin repeat-likeSuperfamily: Glycogen synthesis protein GlgSFamily: Glycogen synthesis protein GlgS2 c1rrzA_
100.0
100
PDB header: structural genomics,biosynthetic proteinChain: A: PDB Molecule: glycogen synthesis protein glgs;PDBTitle: solution structure of glgs protein from e. coli
3 d1se7a_
96.1
31
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like4 d2ae9a1
96.0
27
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like5 d2idob1
95.7
31
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like6 c2azpA_
21.3
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa1268;PDBTitle: crystal structure of pa1268 solved by sulfur sad
7 d1em8a_
17.8
14
Fold: DNA polymerase III chi subunitSuperfamily: DNA polymerase III chi subunitFamily: DNA polymerase III chi subunit8 c2qjxA_
16.2
20
PDB header: protein bindingChain: A: PDB Molecule: protein bim1;PDBTitle: structural basis of microtubule plus end tracking by2 xmap215, clip-170 and eb1
9 c1w62B_
14.1
24
PDB header: racemaseChain: B: PDB Molecule: b-cell mitogen;PDBTitle: proline racemase in complex with one molecule of pyrrole-2-2 carboxylic acid (hemi form)
10 c1hf0A_
13.2
38
PDB header: transcription factorChain: A: PDB Molecule: octamer-binding transcription factor 1;PDBTitle: crystal structure of the dna-binding domain of oct-1 bound2 to dna as a dimer
11 d1v5ka_
9.8
29
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain12 c3d1nK_
9.5
44
PDB header: transcription regulator/dnaChain: K: PDB Molecule: pou domain, class 6, transcription factor 1;PDBTitle: structure of human brn-5 transcription factor in complex2 with corticotrophin-releasing hormone gene promoter
13 d1skva_
9.2
30
Fold: ROP-likeSuperfamily: Hypothetical protein D-63Family: Hypothetical protein D-6314 c1b9uA_
9.1
50
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
15 d1y7ua1
8.6
17
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like16 c2diuA_
8.4
26
PDB header: rna binding proteinChain: A: PDB Molecule: kiaa0430 protein;PDBTitle: solution structure of the rrm domain of kiaa0430 protein
17 d1s6la1
8.0
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like18 c2gm2A_
7.7
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
19 d1au7a1
7.6
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain20 d2qjza1
6.9
24
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain21 d1bama_
not modelled
6.9
33
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BamHI22 d3d1ma1
not modelled
6.8
70
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: Hedgehog (development protein), N-terminal signaling domain23 c3swfA_
not modelled
6.6
44
PDB header: transport proteinChain: A: PDB Molecule: cgmp-gated cation channel alpha-1;PDBTitle: cnga1 621-690 containing clz domain
24 c2hl7A_
not modelled
6.6
40
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
25 c3m1nB_
not modelled
6.6
70
PDB header: signaling proteinChain: B: PDB Molecule: sonic hedgehog protein;PDBTitle: crystal structure of human sonic hedgehog n-terminal domain
26 c1geaA_
not modelled
6.5
42
PDB header: neuropeptideChain: A: PDB Molecule: pituitary adenylate cyclase activatingPDBTitle: receptor-bound conformation of pacap21
27 d1b79a_
not modelled
6.3
24
Fold: N-terminal domain of DnaB helicaseSuperfamily: N-terminal domain of DnaB helicaseFamily: N-terminal domain of DnaB helicase28 d2pp6a1
not modelled
6.1
32
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: gpFII-like29 c3byaB_
not modelled
6.1
71
PDB header: metal binding proteinChain: B: PDB Molecule: glutamate [nmda] receptor subunit zeta-1 peptide;PDBTitle: structure of a calmodulin complex
30 d1b8ia_
not modelled
6.0
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain31 d1ejxb_
not modelled
6.0
39
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit32 c2jtxA_
not modelled
5.9
19
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation factor iie subunitPDBTitle: nmr structure of the tfiie-alpha carboxyl terminus
33 c2yqeA_
not modelled
5.5
15
PDB header: dna binding proteinChain: A: PDB Molecule: jumonji/arid domain-containing protein 1d;PDBTitle: solution structure of the arid domain of jarid1d protein
34 c2rnqA_
not modelled
5.5
19
PDB header: transcriptionChain: A: PDB Molecule: transcription initiation factor iie subunitPDBTitle: solution structure of the c-terminal acidic domain of tfiie2 alpha
35 c2hqwB_
not modelled
5.4
71
PDB header: metal binding proteinChain: B: PDB Molecule: glutamate nmda receptor subunit zeta 1;PDBTitle: crystal structure of ca2+/calmodulin bound to nmda receptor nr1c12 peptide
36 c2kw0A_
not modelled
5.3
33
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli