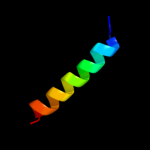
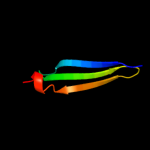
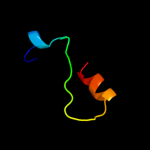
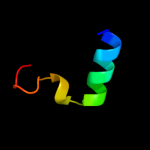
1 d1xmec1
32.1
30
Fold: Single transmembrane helixSuperfamily: Bacterial ba3 type cytochrome c oxidase subunit IIaFamily: Bacterial ba3 type cytochrome c oxidase subunit IIa2 d1im3d_
29.1
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytomegalovirus protein US23 d1e50b_
19.9
26
Fold: Core binding factor beta, CBFSuperfamily: Core binding factor beta, CBFFamily: Core binding factor beta, CBF4 c3ghfA_
19.9
17
PDB header: cell cycleChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the septum site-determining protein2 minc from salmonella typhimurium
5 d2isya2
18.5
17
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain6 c2im5C_
18.0
18
PDB header: transferaseChain: C: PDB Molecule: nicotinate phosphoribosyltransferase;PDBTitle: crystal structure of nicotinate phosphoribosyltransferase2 from porphyromonas gingivalis
7 d2nwaa1
16.4
44
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YtmB-like8 d1gl4a1
14.6
18
Fold: GFP-likeSuperfamily: GFP-likeFamily: Domain G2 of nidogen-19 d1b26a2
11.8
13
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases10 c2cg5A_
11.4
12
PDB header: transferase/hydrolaseChain: A: PDB Molecule: l-aminoadipate-semialdehyde dehydrogenase-PDBTitle: structure of aminoadipate-semialdehyde dehydrogenase-2 phosphopantetheinyl transferase in complex with cytosolic3 acyl carrier protein and coenzyme a
11 c3db3A_
11.2
25
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase uhrf1;PDBTitle: crystal structure of the tandem tudor domains of the e3 ubiquitin-2 protein ligase uhrf1 in complex with trimethylated histone h3-k93 peptide
12 c1h4uA_
8.7
18
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
13 d1v9la2
8.3
14
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases14 d1hwxa2
8.1
14
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases15 d1bgva2
8.0
16
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases16 c1piqA_
8.0
18
PDB header: dna binding proteinChain: A: PDB Molecule: protein (general control protein gcn4-piq);PDBTitle: crystal structure of gcn4-piq, a trimeric coiled coil with buried2 polar residues
17 d1jb0j_
7.3
17
Fold: Single transmembrane helixSuperfamily: Subunit IX of photosystem I reaction centre, PsaJFamily: Subunit IX of photosystem I reaction centre, PsaJ18 c2o01J_
7.3
17
PDB header: photosynthesisChain: J: PDB Molecule: photosystem i reaction center subunit ix;PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
19 d1euza2
7.1
13
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases20 d1xo8a_
6.9
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: LEA14-likeFamily: LEA14-like21 d1bvua2
not modelled
6.7
9
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases22 c1v9lA_
not modelled
6.2
14
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: l-glutamate dehydrogenase from pyrobaculum islandicum2 complexed with nad
23 c2e6gI_
not modelled
6.2
32
PDB header: hydrolaseChain: I: PDB Molecule: 5'-nucleotidase sure;PDBTitle: crystal structure of the stationary phase survival protein sure from2 thermus thermophilus hb8 in complex with phosphate
24 d1ikta_
not modelled
6.1
16
Fold: SCP-likeSuperfamily: SCP-likeFamily: Sterol carrier protein, SCP25 c2wpzA_
not modelled
6.1
16
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with two vxxnxxx motifs2 coordinating chloride
26 d1g3wa2
not modelled
5.8
17
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain27 c2wpzB_
not modelled
5.8
16
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with two vxxnxxx motifs2 coordinating chloride
28 c2wpzC_
not modelled
5.6
16
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with two vxxnxxx motifs2 coordinating chloride
29 d1l5xa_
not modelled
5.6
12
Fold: SurE-likeSuperfamily: SurE-likeFamily: SurE-like30 c3aogA_
not modelled
5.6
13
PDB header: oxidoreductaseChain: A: PDB Molecule: glutamate dehydrogenase;PDBTitle: crystal structure of glutamate dehydrogenase (gdhb) from thermus2 thermophilus (glu bound form)
31 c2ccfA_
not modelled
5.3
24
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel configuration of pli e20s
32 d1gtma2
not modelled
5.3
12
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases33 c1unyB_
not modelled
5.2
24
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
34 c1gcmA_
not modelled
5.1
18
PDB header: transcription regulationChain: A: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
35 c1uo0B_
not modelled
5.1
24
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
36 c1uo0A_
not modelled
5.1
24
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
37 c1uo1B_
not modelled
5.1
24
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
38 c1uo1A_
not modelled
5.1
24
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
39 c1unwB_
not modelled
5.1
24
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
40 c1gclD_
not modelled
5.0
24
PDB header: leucine zipperChain: D: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
41 c1gclB_
not modelled
5.0
24
PDB header: leucine zipperChain: B: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
42 c1gclC_
not modelled
5.0
24
PDB header: leucine zipperChain: C: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
43 c1gclA_
not modelled
5.0
24
PDB header: leucine zipperChain: A: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li