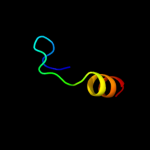
1 d2hlya1
35.4
33
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Atu2299-like2 c3qnqD_
35.0
14
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
3 c3pjzA_
34.3
14
PDB header: transport proteinChain: A: PDB Molecule: potassium uptake protein trkh;PDBTitle: crystal structure of the potassium transporter trkh from vibrio2 parahaemolyticus
4 d1j4na_
21.8
22
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like5 c3ajvA_
17.3
16
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: splicing endonuclease from aeropyrum pernix
6 c2ntxB_
16.3
29
PDB header: signaling proteinChain: B: PDB Molecule: emb7 c3ai9X_
16.0
42
PDB header: transferaseChain: X: PDB Molecule: upf0217 protein mj1640;PDBTitle: crystal structure of duf358 protein reveals a putative spout-class2 rrna methyltransferase
8 c2ka2A_
15.2
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
9 c2ka2B_
15.2
33
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
10 c2ka1B_
15.2
33
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
11 c2ka1A_
15.2
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
12 d2qwva1
15.0
25
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF1056-like13 d2qmma1
13.6
27
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF1056-like14 d1acca_
13.1
16
Fold: Anthrax protective antigenSuperfamily: Anthrax protective antigenFamily: Anthrax protective antigen15 d1ig8a1
11.7
60
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase16 c2j5dA_
11.2
32
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
17 d1fx8a_
11.1
20
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like18 c1ldaA_
11.1
20
PDB header: transport proteinChain: A: PDB Molecule: glycerol uptake facilitator protein;PDBTitle: crystal structure of the e. coli glycerol facilitator (glpf) without2 substrate glycerol
19 c3zy6A_
11.1
29
PDB header: transferaseChain: A: PDB Molecule: putative gdp-fucose protein o-fucosyltransferase 1;PDBTitle: crystal structure of pofut1 in complex with gdp-fucose2 (crystal-form-ii)
20 d1czan1
10.8
50
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase21 c2b2hA_
not modelled
10.2
26
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter;PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
22 c3op9A_
not modelled
10.0
14
PDB header: transcription regulatorChain: A: PDB Molecule: pli0006 protein;PDBTitle: crystal structure of transcriptional regulator from listeria innocua
23 d1czan3
not modelled
9.9
60
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase24 d1bdga1
not modelled
9.3
60
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase25 d1bg3a3
not modelled
9.2
60
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase26 d1bg3a1
not modelled
9.1
50
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase27 c2k3uA_
not modelled
8.8
37
PDB header: immune systemChain: A: PDB Molecule: chemotaxis inhibitory protein;PDBTitle: structure of the tyrosine-sulfated c5a receptor n-terminus2 in complex with the immune evasion protein chips.
28 c3llqB_
not modelled
8.8
19
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin z 2;PDBTitle: aquaporin structure from plant pathogen agrobacterium tumerfaciens
29 d1v4sa1
not modelled
8.4
50
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase30 c2lmfA_
not modelled
7.2
43
PDB header: antimicrobial proteinChain: A: PDB Molecule: antibacterial protein ll-37;PDBTitle: solution structure of human ll-23 bound to membrane-mimetic micelles
31 c3hm8D_
not modelled
7.0
60
PDB header: transferaseChain: D: PDB Molecule: hexokinase-3;PDBTitle: crystal structure of the c-terminal hexokinase domain of human hk3
32 c2w2eA_
not modelled
6.9
26
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin;PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin,2 aqy1, in a closed conformation at ph 3.5
33 d1rc2a_
not modelled
6.9
13
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like34 c3p04B_
not modelled
6.6
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized bcr;PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
35 c2magA_
not modelled
6.4
35
PDB header: antibioticChain: A: PDB Molecule: magainin 2;PDBTitle: nmr structure of magainin 2 in dpc micelles, 10 structures
36 c3c9jD_
not modelled
6.2
21
PDB header: membrane proteinChain: D: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
37 c3c9jA_
not modelled
6.2
21
PDB header: membrane proteinChain: A: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
38 c3c9jB_
not modelled
6.2
21
PDB header: membrane proteinChain: B: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
39 c3c9jC_
not modelled
6.2
21
PDB header: membrane proteinChain: C: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
40 c3ct5A_
not modelled
6.0
14
PDB header: hydrolaseChain: A: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall degrading enzyme2 in the bacteriophage phi29 tail
41 d1u9ya2
not modelled
6.0
38
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like42 d1u7ga_
not modelled
5.9
16
Fold: Ammonium transporterSuperfamily: Ammonium transporterFamily: Ammonium transporter43 d1wi9a_
not modelled
5.7
63
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PCI domain (PINT motif)44 c1ymgA_
not modelled
5.7
26
PDB header: membrane proteinChain: A: PDB Molecule: lens fiber major intrinsic protein;PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
45 d1ymga1
not modelled
5.7
26
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like46 d1l8wd_
not modelled
5.6
35
Fold: Variable surface antigen VlsESuperfamily: Variable surface antigen VlsEFamily: Variable surface antigen VlsE47 c3c1iA_
not modelled
5.5
20
PDB header: transport proteinChain: A: PDB Molecule: ammonia channel;PDBTitle: substrate binding, deprotonation and selectivity at the2 periplasmic entrance of the e. coli ammonia channel amtb
48 c1iyjC_
not modelled
5.5
55
PDB header: gene regulation/antitumor proteinChain: C: PDB Molecule: deleted in split hand/split foot protein 1;PDBTitle: structure of a brca2-dss1 complex
49 c1bdgA_
not modelled
5.5
60
PDB header: hexokinaseChain: A: PDB Molecule: hexokinase;PDBTitle: hexokinase from schistosoma mansoni complexed with glucose
50 c1v4sA_
not modelled
5.5
50
PDB header: transferaseChain: A: PDB Molecule: glucokinase isoform 2;PDBTitle: crystal structure of human glucokinase
51 c1ig8A_
not modelled
5.4
60
PDB header: transferaseChain: A: PDB Molecule: hexokinase pii;PDBTitle: crystal structure of yeast hexokinase pii with the correct2 amino acid sequence
52 c2oarA_
not modelled
5.3
26
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
53 c3hzqA_
not modelled
5.3
26
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
54 c3pvpA_
not modelled
5.1
10
PDB header: dna binding protein/dnaChain: A: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: structure of mycobacterium tuberculosis dnaa-dbd in complex with box22 dna