| 1 |
|
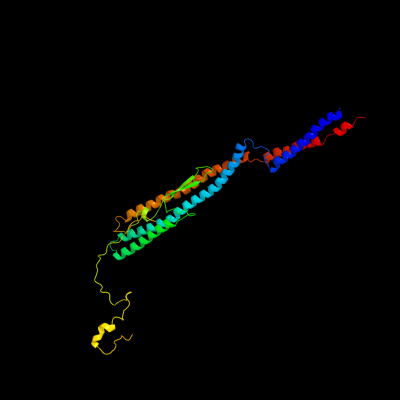
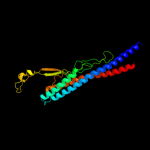
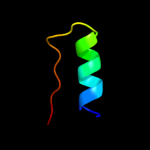
PDB 1ucu chain A
Region: 1 - 317
Aligned: 317
Modelled: 317
Confidence: 100.0%
Identity: 21%
Fold: Phase 1 flagellin
Superfamily: Phase 1 flagellin
Family: Phase 1 flagellin
Phyre2
| 2 |
|
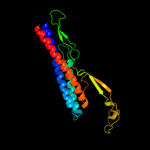
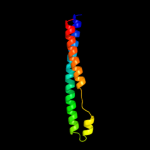
PDB 3k8v chain B
Region: 44 - 300
Aligned: 257
Modelled: 257
Confidence: 100.0%
Identity: 19%
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
Phyre2
| 3 |
|
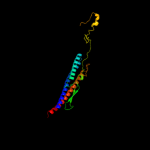
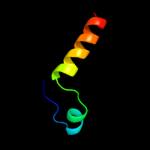
PDB 2d4x chain A
Region: 50 - 262
Aligned: 213
Modelled: 213
Confidence: 100.0%
Identity: 81%
PDB header:structural protein
Chain: A: PDB Molecule:flagellar hook-associated protein 3;
PDBTitle: crystal structure of a 26k fragment of hap3 (flgl)
Phyre2
| 4 |
|
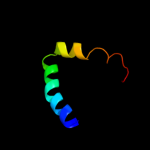
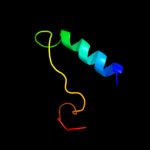
PDB 3k8w chain A
Region: 42 - 275
Aligned: 234
Modelled: 234
Confidence: 100.0%
Identity: 19%
PDB header:structural protein
Chain: A: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c45
Phyre2
| 5 |
|
PDB 3pwx chain B
Region: 42 - 278
Aligned: 194
Modelled: 201
Confidence: 100.0%
Identity: 24%
PDB header:structural protein
Chain: B: PDB Molecule:putative flagellar hook-associated protein;
PDBTitle: structure of putative flagellar hook-associated protein from vibrio2 parahaemolyticus
Phyre2
| 6 |
|
PDB 2zbi chain B
Region: 61 - 264
Aligned: 204
Modelled: 204
Confidence: 100.0%
Identity: 22%
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin
Phyre2
| 7 |
|
PDB 1io1 chain A
Region: 56 - 275
Aligned: 220
Modelled: 220
Confidence: 100.0%
Identity: 22%
Fold: Phase 1 flagellin
Superfamily: Phase 1 flagellin
Family: Phase 1 flagellin
Phyre2
| 8 |
|
PDB 1ory chain B
Region: 280 - 317
Aligned: 38
Modelled: 37
Confidence: 94.1%
Identity: 18%
PDB header:chaperone
Chain: B: PDB Molecule:flagellin;
PDBTitle: flagellar export chaperone in complex with its cognate binding partner
Phyre2
| 9 |
|
PDB 3a69 chain A
Region: 280 - 310
Aligned: 31
Modelled: 31
Confidence: 45.8%
Identity: 10%
PDB header:motor protein
Chain: A: PDB Molecule:flagellar hook protein flge;
PDBTitle: atomic model of the bacterial flagellar hook based on2 docking an x-ray derived structure and terminal two alpha-3 helices into an 7.1 angstrom resolution cryoem map
Phyre2
| 10 |
|
PDB 2kyy chain A
Region: 115 - 137
Aligned: 23
Modelled: 23
Confidence: 9.3%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:possible atp-dependent dna helicase recg-related protein;
PDBTitle: solution nmr structure of the n-terminal domain of putative atp-2 dependent dna helicase recg-related protein from nitrosomonas3 europaea, northeast structural genomics consortium target ner70a
Phyre2
| 11 |
|
PDB 2ieq chain C
Region: 229 - 310
Aligned: 76
Modelled: 82
Confidence: 8.5%
Identity: 12%
PDB header:viral protein
Chain: C: PDB Molecule:spike glycoprotein;
PDBTitle: core structure of s2 from the human coronavirus nl63 spike2 glycoprotein
Phyre2
| 12 |
|
PDB 3pzd chain B
Region: 37 - 67
Aligned: 31
Modelled: 31
Confidence: 8.3%
Identity: 19%
PDB header:motor protein/apoptosis
Chain: B: PDB Molecule:netrin receptor dcc;
PDBTitle: structure of the myosin x myth4-ferm/dcc complex
Phyre2
| 13 |
|
PDB 2do9 chain A
Region: 281 - 316
Aligned: 36
Modelled: 36
Confidence: 7.0%
Identity: 19%
PDB header:signaling protein
Chain: A: PDB Molecule:nacht-, lrr- and pyd-containing protein 10;
PDBTitle: solution structure of the pyrin/paad-dapin domain in mouse2 nalp10 (nacht, leucine rich repeat and pyd containing 10)
Phyre2
| 14 |
|
PDB 1ffv chain A domain 2
Region: 309 - 317
Aligned: 9
Modelled: 9
Confidence: 7.0%
Identity: 11%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: 2Fe-2S ferredoxin-like
Family: 2Fe-2S ferredoxin domains from multidomain proteins
Phyre2
| 15 |
|
PDB 3lmm chain A
Region: 112 - 146
Aligned: 34
Modelled: 35
Confidence: 6.7%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the dip2311 protein from2 corynebacterium diphtheriae, northeast structural genomics3 consortium target cdr35
Phyre2
| 16 |
|
PDB 3a1y chain F
Region: 271 - 289
Aligned: 19
Modelled: 19
Confidence: 6.4%
Identity: 16%
PDB header:ribosomal protein
Chain: F: PDB Molecule:50s ribosomal protein p1 (l12p);
PDBTitle: the structure of protein complex
Phyre2
| 17 |
|
PDB 2w6j chain G
Region: 243 - 290
Aligned: 48
Modelled: 48
Confidence: 6.2%
Identity: 6%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: low resolution structures of bovine mitochondrial f1-atpase2 during controlled dehydration: hydration state 5.
Phyre2
| 18 |
|
PDB 3fks chain Y
Region: 243 - 290
Aligned: 48
Modelled: 48
Confidence: 6.2%
Identity: 8%
PDB header:hydrolase
Chain: Y: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: yeast f1 atpase in the absence of bound nucleotides
Phyre2