| 1 |
|
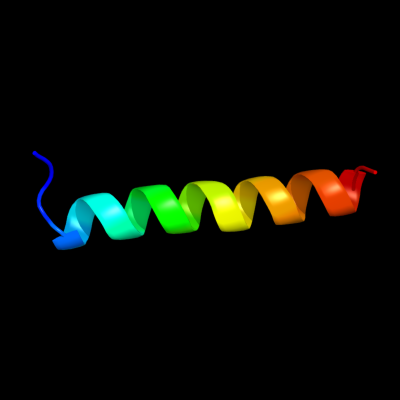
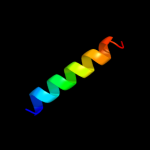
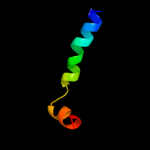
PDB 2wl8 chain D
Region: 71 - 95
Aligned: 25
Modelled: 25
Confidence: 25.0%
Identity: 12%
PDB header:protein transport
Chain: D: PDB Molecule:peroxisomal biogenesis factor 19;
PDBTitle: x-ray crystal structure of pex19p
Phyre2
| 2 |
|
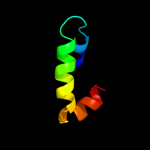
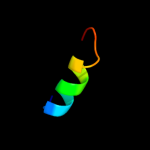
PDB 1q2z chain A
Region: 61 - 98
Aligned: 38
Modelled: 38
Confidence: 18.6%
Identity: 11%
Fold: alpha-alpha superhelix
Superfamily: C-terminal domain of Ku80
Family: C-terminal domain of Ku80
Phyre2
| 3 |
|
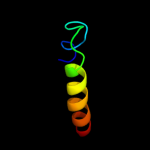
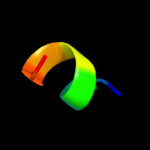
PDB 2jee chain A
Region: 104 - 114
Aligned: 11
Modelled: 11
Confidence: 15.0%
Identity: 36%
PDB header:cell cycle
Chain: A: PDB Molecule:yiiu;
PDBTitle: xray structure of e. coli yiiu
Phyre2
| 4 |
|
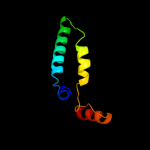
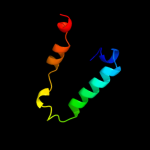
PDB 1faf chain A
Region: 68 - 98
Aligned: 30
Modelled: 31
Confidence: 14.6%
Identity: 7%
Fold: Long alpha-hairpin
Superfamily: Chaperone J-domain
Family: Chaperone J-domain
Phyre2
| 5 |
|
PDB 2pi8 chain A domain 1
Region: 59 - 89
Aligned: 31
Modelled: 31
Confidence: 13.5%
Identity: 19%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 6 |
|
PDB 2cwo chain D
Region: 48 - 121
Aligned: 67
Modelled: 74
Confidence: 12.4%
Identity: 19%
PDB header:rna binding protein
Chain: D: PDB Molecule:rna silencing suppressor;
PDBTitle: crystal structure of rna silencing suppressor p21 from beet yellows2 virus
Phyre2
| 7 |
|
PDB 3dve chain B
Region: 90 - 102
Aligned: 13
Modelled: 13
Confidence: 11.0%
Identity: 23%
PDB header:membrane protein
Chain: B: PDB Molecule:voltage-dependent n-type calcium channel subunit alpha-1b;
PDBTitle: crystal structure of ca2+/cam-cav2.2 iq domain complex
Phyre2
| 8 |
|
PDB 2pjw chain V
Region: 76 - 96
Aligned: 21
Modelled: 21
Confidence: 8.7%
Identity: 33%
PDB header:endocytosis/exocytosis
Chain: V: PDB Molecule:vacuolar protein sorting-associated protein 27;
PDBTitle: the vps27/hse1 complex is a gat domain-based scaffold for2 ubiquitin-dependent sorting
Phyre2
| 9 |
|
PDB 1k3v chain A
Region: 72 - 87
Aligned: 16
Modelled: 16
Confidence: 8.3%
Identity: 19%
PDB header:virus
Chain: A: PDB Molecule:capsid protein vp2;
PDBTitle: porcine parvovirus capsid
Phyre2
| 10 |
|
PDB 1k3v chain A
Region: 72 - 87
Aligned: 16
Modelled: 16
Confidence: 8.3%
Identity: 19%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: ssDNA viruses
Family: Parvoviridae-like VP
Phyre2
| 11 |
|
PDB 1c8d chain A
Region: 72 - 87
Aligned: 16
Modelled: 16
Confidence: 7.7%
Identity: 13%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: ssDNA viruses
Family: Parvoviridae-like VP
Phyre2
| 12 |
|
PDB 2wx4 chain C
Region: 16 - 23
Aligned: 8
Modelled: 8
Confidence: 7.4%
Identity: 50%
PDB header:structural protein
Chain: C: PDB Molecule:decapping protein 1;
PDBTitle: asymmetric trimer of the drosophila melanogaster dcp1 c-2 terminal domain
Phyre2
| 13 |
|
PDB 1kl1 chain A
Region: 66 - 116
Aligned: 50
Modelled: 51
Confidence: 7.1%
Identity: 16%
Fold: PLP-dependent transferase-like
Superfamily: PLP-dependent transferases
Family: GABA-aminotransferase-like
Phyre2
| 14 |
|
PDB 2nw8 chain A domain 1
Region: 63 - 82
Aligned: 20
Modelled: 20
Confidence: 7.1%
Identity: 20%
Fold: Indolic compounds 2,3-dioxygenase-like
Superfamily: Indolic compounds 2,3-dioxygenase-like
Family: Bacterial tryptophan 2,3-dioxygenase
Phyre2
| 15 |
|
PDB 2nox chain P
Region: 55 - 82
Aligned: 28
Modelled: 28
Confidence: 6.9%
Identity: 21%
PDB header:oxidoreductase
Chain: P: PDB Molecule:tryptophan 2,3-dioxygenase;
PDBTitle: crystal structure of tryptophan 2,3-dioxygenase from ralstonia2 metallidurans
Phyre2
| 16 |
|
PDB 2elb chain A domain 1
Region: 25 - 118
Aligned: 94
Modelled: 94
Confidence: 6.8%
Identity: 7%
Fold: BAR/IMD domain-like
Superfamily: BAR/IMD domain-like
Family: BAR domain
Phyre2
| 17 |
|
PDB 1s58 chain A
Region: 72 - 87
Aligned: 16
Modelled: 16
Confidence: 6.3%
Identity: 25%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: ssDNA viruses
Family: Parvoviridae-like VP
Phyre2
| 18 |
|
PDB 3dvj chain B
Region: 90 - 99
Aligned: 10
Modelled: 10
Confidence: 5.8%
Identity: 30%
PDB header:membrane protein
Chain: B: PDB Molecule:voltage-dependent n-type calcium channel subunit alpha-1b;
PDBTitle: crystal structure of ca2+/cam-cav2.2 iq domain (without cloning2 artifact, hm to tv) complex
Phyre2
| 19 |
|
PDB 2nw7 chain C
Region: 63 - 82
Aligned: 20
Modelled: 20
Confidence: 5.7%
Identity: 20%
PDB header:oxidoreductase
Chain: C: PDB Molecule:tryptophan 2,3-dioxygenase;
PDBTitle: crystal structure of tryptophan 2,3-dioxygenase (tdo) from2 xanthomonas campestris in complex with ferric heme.3 northeast structural genomics target xcr13
Phyre2
| 20 |
|
PDB 1fpv chain A
Region: 72 - 87
Aligned: 16
Modelled: 16
Confidence: 5.7%
Identity: 13%
PDB header:virus
Chain: A: PDB Molecule:feline panleukopenia virus (strain b) viral
PDBTitle: structure determination of feline panleukopenia virus empty2 particles
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|