| 1 | d1gmea_ |
|
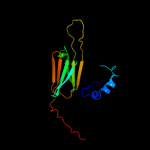 |
99.9 |
22 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
| 2 | d1gmeb_ |
|
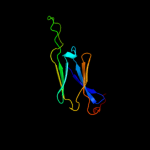 |
99.9 |
23 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
| 3 | d1shsa_ |
|
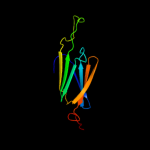 |
99.9 |
23 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
| 4 | c1shsD_ |
|
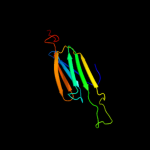 |
99.9 |
23 |
PDB header:heat shock protein
Chain: D: PDB Molecule:small heat shock protein;
PDBTitle: small heat shock protein from methanococcus jannaschii
|
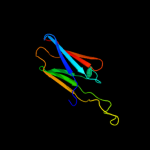
| 5 | c3glaA_ |
|
 |
99.8 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:low molecular weight heat shock protein;
PDBTitle: crystal structure of the hspa from xanthomonas axonopodis
|
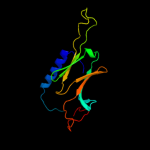
| 6 | c2bolA_ |
|
 |
99.8 |
22 |
PDB header:heat shock protein
Chain: A: PDB Molecule:small heat shock protein;
PDBTitle: crystal structure and assembly of tsp36, a metazoan small2 heat shock protein
|
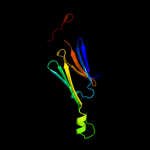
| 7 | c3l1eA_ |
|
 |
99.8 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:alpha-crystallin a chain;
PDBTitle: bovine alphaa crystallin zinc bound
|
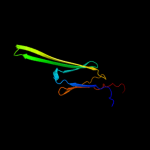
| 8 | c2wj5A_ |
|
 |
99.8 |
17 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein beta-6;
PDBTitle: rat alpha crystallin domain
|
| 9 | c3aabA_ |
|
 |
99.8 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:putative uncharacterized protein st1653;
PDBTitle: small heat shock protein hsp14.0 with the mutations of i120f and i122f2 in the form i crystal
|
| 10 | d2h50a1 |
|
 |
99.7 |
24 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
| 11 | c2klrA_ |
|
 |
99.7 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:alpha-crystallin b chain;
PDBTitle: solid-state nmr structure of the alpha-crystallin domain in alphab-2 crystallin oligomers
|
| 12 | c3q9qB_ |
|
 |
99.7 |
15 |
PDB header:chaperone
Chain: B: PDB Molecule:heat shock protein beta-1;
PDBTitle: hspb1 fragment second crystal form
|
| 13 | c2wj7D_ |
|
 |
99.5 |
18 |
PDB header:chaperone
Chain: D: PDB Molecule:alpha-crystallin b chain;
PDBTitle: human alphab crystallin
|
| 14 | c3igfB_ |
|
 |
98.0 |
25 |
PDB header:atp binding protein
Chain: B: PDB Molecule:all4481 protein;
PDBTitle: crystal structure of the all4481 protein from nostoc sp. pcc 7120,2 northeast structural genomics consortium target nsr300
|
| 15 | d1rl1a_ |
|
 |
97.8 |
16 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:GS domain |
| 16 | c2jkiS_ |
|
 |
97.8 |
13 |
PDB header:chaperone
Chain: S: PDB Molecule:sgt1-like protein;
PDBTitle: complex of hsp90 n-terminal and sgt1 cs domain
|
| 17 | c3eudE_ |
|
 |
96.8 |
16 |
PDB header:nuclear protein
Chain: E: PDB Molecule:protein shq1;
PDBTitle: structure of the cs domain of the essential h/aca rnp2 assembly protein shq1p
|
| 18 | d1wh0a_ |
|
 |
96.7 |
8 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:GS domain |
| 19 | c1x5mA_ |
|
 |
96.4 |
16 |
PDB header:apoptosis, signaling protein
Chain: A: PDB Molecule:calcyclin-binding protein;
PDBTitle: solution structure of the core domain of calcyclin binding2 protein; siah-interacting protein (sip)
|
| 20 | d1ejfa_ |
|
 |
96.3 |
10 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:Co-chaperone p23-like |
| 21 | d1wfia_ |
|
not modelled |
96.0 |
13 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:Nuclear movement domain |
| 22 | d1wgva_ |
|
not modelled |
95.8 |
7 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:Nuclear movement domain |
| 23 | c2k8qA_ |
|
not modelled |
95.5 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:protein shq1;
PDBTitle: nmr structure of shq1p n-terminal domain
|
| 24 | d1xo9a_ |
|
not modelled |
95.1 |
19 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:Co-chaperone p23-like |
| 25 | c2o30A_ |
|
not modelled |
93.8 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nuclear movement protein;
PDBTitle: nuclear movement protein from e. cuniculi gb-m1
|
| 26 | c2rh0B_ |
|
not modelled |
92.2 |
9 |
PDB header:nuclear protein
Chain: B: PDB Molecule:nudc domain-containing protein 2;
PDBTitle: crystal structure of nudc domain-containing protein 22 (13542905) from mus musculus at 1.95 a resolution
|
| 27 | c2cg9Y_ |
|
not modelled |
70.5 |
14 |
PDB header:chaperone
Chain: Y: PDB Molecule:co-chaperone protein sba1;
PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
|
| 28 | d2fqla1 |
|
not modelled |
65.2 |
11 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 29 | d2ga5a1 |
|
not modelled |
65.0 |
11 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 30 | d1ew4a_ |
|
not modelled |
50.8 |
22 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 31 | d2cqla1 |
|
not modelled |
47.4 |
30 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 32 | d1ekga_ |
|
not modelled |
46.2 |
19 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 33 | d1vqoe1 |
|
not modelled |
38.2 |
24 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 34 | d1rl6a1 |
|
not modelled |
37.1 |
24 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 35 | d2j01h1 |
|
not modelled |
31.9 |
24 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 36 | c4a1eE_ |
|
not modelled |
29.3 |
24 |
PDB header:ribosome
Chain: E: PDB Molecule:60s ribosomal protein l9;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
|
| 37 | c3iz5F_ |
|
not modelled |
28.7 |
24 |
PDB header:ribosome
Chain: F: PDB Molecule:60s ribosomal protein l9 (l6p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 38 | c2zkre_ |
|
not modelled |
28.7 |
41 |
PDB header:ribosomal protein/rna
Chain: E: PDB Molecule:rna expansion segment es7 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 39 | c3ccmE_ |
|
not modelled |
24.3 |
24 |
PDB header:ribosome
Chain: E: PDB Molecule:50s ribosomal protein l6p;
PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation g2611u
|
| 40 | d1h8la1 |
|
not modelled |
21.9 |
12 |
Fold:Prealbumin-like
Superfamily:Carboxypeptidase regulatory domain-like
Family:Carboxypeptidase regulatory domain |
| 41 | c3ho6B_ |
|
not modelled |
19.1 |
16 |
PDB header:toxin
Chain: B: PDB Molecule:toxin a;
PDBTitle: structure-function analysis of inositol hexakisphosphate-2 induced autoprocessing in clostridium difficile toxin a
|
| 42 | c3muwU_ |
|
not modelled |
18.9 |
10 |
PDB header:virus
Chain: U: PDB Molecule:structural polyprotein;
PDBTitle: pseudo-atomic structure of the e2-e1 protein shell in sindbis virus
|
| 43 | c2hguH_ |
|
not modelled |
18.5 |
24 |
PDB header:ribosome
Chain: H: PDB Molecule:50s ribosomal protein l6;
PDBTitle: 70s t.th. ribosome functional complex with mrna and e- and p-site2 trnas at 4.5a. this entry 2hgu contains 50s ribosomal subunit. the3 30s ribosomal subunit can be found in pdb entry 2hgr.
|
| 44 | c2xfbI_ |
|
not modelled |
16.7 |
20 |
PDB header:virus
Chain: I: PDB Molecule:e2 envelope glycoprotein;
PDBTitle: the chikungunya e1 e2 envelope glycoprotein complex fit into2 the sindbis virus cryo-em map
|
| 45 | c3n43B_ |
|
not modelled |
16.6 |
20 |
PDB header:viral protein
Chain: B: PDB Molecule:e2 envelope glycoprotein;
PDBTitle: crystal structures of the mature envelope glycoprotein complex2 (trypsin cleavage) of chikungunya virus.
|
| 46 | c3petA_ |
|
not modelled |
16.0 |
8 |
PDB header:cell adhesion
Chain: A: PDB Molecule:putative adhesin;
PDBTitle: crystal structure of a putative adhesin (bf0245) from bacteroides2 fragilis nctc 9343 at 2.07 a resolution
|
| 47 | c3n40P_ |
|
not modelled |
15.5 |
20 |
PDB header:viral protein
Chain: P: PDB Molecule:p62 envelope glycoprotein;
PDBTitle: crystal structure of the immature envelope glycoprotein complex of2 chikungunya virus.
|
| 48 | c3ipfA_ |
|
not modelled |
15.1 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the q251q8_deshy protein from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr8c.
|
| 49 | c2gjhA_ |
|
not modelled |
13.7 |
26 |
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein;
PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
|
| 50 | d2zjre2 |
|
not modelled |
13.4 |
18 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 51 | c487dJ_ |
|
not modelled |
13.2 |
17 |
PDB header:ribosome
Chain: J: PDB Molecule:protein (50s l6 ribosomal protein);
PDBTitle: seven ribosomal proteins fitted to a cryo-electron2 microscopic map of the large 50s subunit at 7.5 angstroms3 resolution
|
| 52 | c3j0cH_ |
|
not modelled |
13.0 |
10 |
PDB header:virus
Chain: H: PDB Molecule:e2 envelope glycoprotein;
PDBTitle: models of e1, e2 and cp of venezuelan equine encephalitis virus tc-832 strain restrained by a near atomic resolution cryo-em map
|
| 53 | c3bboI_ |
|
not modelled |
12.2 |
47 |
PDB header:ribosome
Chain: I: PDB Molecule:ribosomal protein l6;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
|
| 54 | c3ljyA_ |
|
not modelled |
10.8 |
8 |
PDB header:cell adhesion
Chain: A: PDB Molecule:putative adhesin;
PDBTitle: crystal structure of putative adhesin (yp_001304413.1) from2 parabacteroides distasonis atcc 8503 at 2.41 a resolution
|
| 55 | c2wvsD_ |
|
not modelled |
10.5 |
3 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 56 | d1v7wa1 |
|
not modelled |
10.4 |
15 |
Fold:alpha/alpha toroid
Superfamily:Six-hairpin glycosidases
Family:Glycosyltransferase family 36 C-terminal domain |
| 57 | d1uwya1 |
|
not modelled |
10.2 |
13 |
Fold:Prealbumin-like
Superfamily:Carboxypeptidase regulatory domain-like
Family:Carboxypeptidase regulatory domain |
| 58 | c2lezA_ |
|
not modelled |
9.8 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:secreted effector protein pipb2;
PDBTitle: solution nmr structure of n-terminal domain of salmonella effector2 protein pipb2. northeast structural genomics consortium (nesg) target3 stt318a
|
| 59 | d2qamg1 |
|
not modelled |
9.8 |
18 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 60 | c2i2vG_ |
|
not modelled |
9.3 |
18 |
PDB header:ribosome
Chain: G: PDB Molecule:50s ribosomal protein l6;
PDBTitle: crystal structure of ribosome with messenger rna and the2 anticodon stem-loop of p-site trna. this file contains the3 50s subunit of one 70s ribosome. the entire crystal4 structure contains two 70s ribosomes and is described in5 remark 400.
|
| 61 | c1sm1E_ |
|
not modelled |
8.7 |
18 |
PDB header:ribosome/antibiotic
Chain: E: PDB Molecule:50s ribosomal protein l6;
PDBTitle: complex of the large ribosomal subunit from deinococcus radiodurans2 with quinupristin and dalfopristin
|
| 62 | c2nsmA_ |
|
not modelled |
8.6 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxypeptidase n catalytic chain;
PDBTitle: crystal structure of the human carboxypeptidase n (kininase i)2 catalytic domain
|
| 63 | c1qysA_ |
|
not modelled |
7.8 |
24 |
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
|
| 64 | c3pa8A_ |
|
not modelled |
7.5 |
11 |
PDB header:toxin/peptide inhibitor
Chain: A: PDB Molecule:toxin b;
PDBTitle: structure of the c. difficile tcdb cysteine protease domain in complex2 with a peptide inhibitor
|
| 65 | c3muuA_ |
|
not modelled |
6.9 |
10 |
PDB header:viral protein
Chain: A: PDB Molecule:structural polyprotein;
PDBTitle: crystal structure of the sindbis virus e2-e1 heterodimer at low ph
|
| 66 | c2x89G_ |
|
not modelled |
6.6 |
10 |
PDB header:immune system
Chain: G: PDB Molecule:beta-2-microglobulin;
PDBTitle: structure of the beta2_microglobulin involved in2 amyloidogenesis
|
| 67 | c1uwyA_ |
|
not modelled |
6.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxypeptidase m;
PDBTitle: crystal structure of human carboxypeptidase m
|
| 68 | d1c12b2 |
|
not modelled |
6.2 |
7 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:C1 set domains (antibody constant domain-like) |
| 69 | d1mvfd_ |
|
not modelled |
6.0 |
20 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:Kis/PemI addiction antidote |
| 70 | d1ji1a2 |
|
not modelled |
6.0 |
17 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
| 71 | c2y9jt_ |
|
not modelled |
5.9 |
13 |
PDB header:protein transport
Chain: T: PDB Molecule:protein prgh;
PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
|
| 72 | d1r3ha1 |
|
not modelled |
5.9 |
7 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:C1 set domains (antibody constant domain-like) |
| 73 | d1pz5b2 |
|
not modelled |
5.7 |
7 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:C1 set domains (antibody constant domain-like) |
| 74 | d1je6a1 |
|
not modelled |
5.5 |
8 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:C1 set domains (antibody constant domain-like) |

