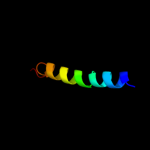| 1 |
|
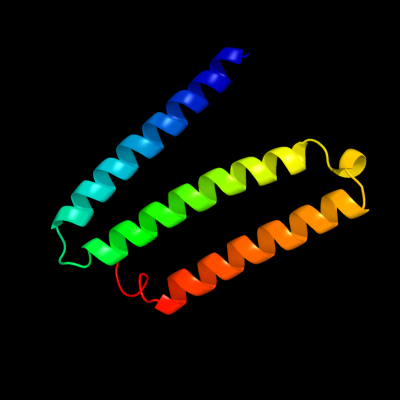
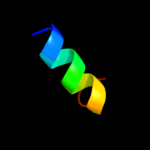
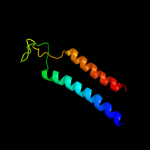
PDB 3rko chain A
Region: 15 - 126
Aligned: 95
Modelled: 95
Confidence: 100.0%
Identity: 98%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh-quinone oxidoreductase subunit a;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
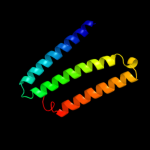
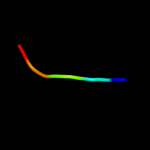
PDB 2k21 chain A
Region: 100 - 137
Aligned: 31
Modelled: 38
Confidence: 32.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2
| 3 |
|
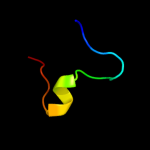
PDB 1jwy chain A domain 1
Region: 41 - 53
Aligned: 13
Modelled: 13
Confidence: 25.8%
Identity: 15%
Fold: SH3-like barrel
Superfamily: Myosin S1 fragment, N-terminal domain
Family: Myosin S1 fragment, N-terminal domain
Phyre2
| 4 |
|
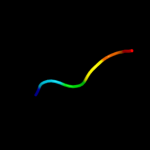
PDB 3bz7 chain A domain 1
Region: 41 - 53
Aligned: 13
Modelled: 13
Confidence: 25.5%
Identity: 15%
Fold: SH3-like barrel
Superfamily: Myosin S1 fragment, N-terminal domain
Family: Myosin S1 fragment, N-terminal domain
Phyre2
| 5 |
|
PDB 1dtd chain B
Region: 119 - 134
Aligned: 16
Modelled: 16
Confidence: 20.2%
Identity: 38%
Fold: Carboxypeptidase inhibitor
Superfamily: Carboxypeptidase inhibitor
Family: Carboxypeptidase inhibitor
Phyre2
| 6 |
|
PDB 3fy6 chain A
Region: 110 - 124
Aligned: 15
Modelled: 15
Confidence: 16.9%
Identity: 40%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:integron cassette protein;
PDBTitle: structure from the mobile metagenome of v. cholerae.2 integron cassette protein vch_cass3
Phyre2
| 7 |
|
PDB 1zrt chain D
Region: 101 - 120
Aligned: 19
Modelled: 20
Confidence: 14.2%
Identity: 21%
PDB header:oxidoreductase/metal transport
Chain: D: PDB Molecule:cytochrome c1;
PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
Phyre2
| 8 |
|
PDB 2fyn chain H
Region: 101 - 120
Aligned: 19
Modelled: 20
Confidence: 11.2%
Identity: 37%
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c1;
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
Phyre2
| 9 |
|
PDB 2csh chain A domain 2
Region: 49 - 53
Aligned: 5
Modelled: 5
Confidence: 10.9%
Identity: 40%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 10 |
|
PDB 1k32 chain A domain 1
Region: 38 - 58
Aligned: 21
Modelled: 21
Confidence: 9.2%
Identity: 38%
Fold: PDZ domain-like
Superfamily: PDZ domain-like
Family: Tail specific protease PDZ domain
Phyre2
| 11 |
|
PDB 2adr chain A domain 2
Region: 49 - 54
Aligned: 6
Modelled: 6
Confidence: 8.6%
Identity: 33%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 12 |
|
PDB 1t0j chain C
Region: 117 - 138
Aligned: 22
Modelled: 12
Confidence: 6.5%
Identity: 18%
PDB header:signaling protein
Chain: C: PDB Molecule:voltage-dependent l-type calcium channel alpha-1c subunit;
PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
Phyre2
| 13 |
|
PDB 2yiu chain E
Region: 101 - 120
Aligned: 19
Modelled: 20
Confidence: 6.2%
Identity: 42%
PDB header:oxidoreductase
Chain: E: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
Phyre2
| 14 |
|
PDB 3mk7 chain F
Region: 12 - 87
Aligned: 75
Modelled: 76
Confidence: 6.1%
Identity: 15%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 15 |
|
PDB 1vyt chain F
Region: 117 - 140
Aligned: 24
Modelled: 14
Confidence: 5.6%
Identity: 17%
PDB header:transport protein
Chain: F: PDB Molecule:voltage-dependent l-type calcium channel
PDBTitle: beta3 subunit complexed with aid
Phyre2
| 16 |
|
PDB 1p84 chain D
Region: 101 - 130
Aligned: 29
Modelled: 30
Confidence: 5.2%
Identity: 17%
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
Phyre2