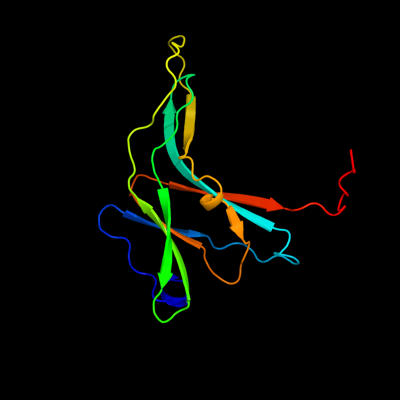
1 d1xxfc_
100.0
96
Fold: Ecotin, trypsin inhibitorSuperfamily: Ecotin, trypsin inhibitorFamily: Ecotin, trypsin inhibitor2 c2y6tE_
100.0
70
PDB header: hydrolase/inhibitorChain: E: PDB Molecule: ecotin;PDBTitle: molecular recognition of chymotrypsin by the serine2 protease inhibitor ecotin from yersinia pestis
3 d1slua_
100.0
94
Fold: Ecotin, trypsin inhibitorSuperfamily: Ecotin, trypsin inhibitorFamily: Ecotin, trypsin inhibitor4 d1ezsa_
100.0
98
Fold: Ecotin, trypsin inhibitorSuperfamily: Ecotin, trypsin inhibitorFamily: Ecotin, trypsin inhibitor5 c1fi8E_
100.0
92
PDB header: hydrolase/hydrolase inhibitorChain: E: PDB Molecule: ecotin;PDBTitle: rat granzyme b [n66q] complexed to ecotin [81-84 iepd]
6 c1fi8F_
99.7
100
PDB header: hydrolase/hydrolase inhibitorChain: F: PDB Molecule: ecotin;PDBTitle: rat granzyme b [n66q] complexed to ecotin [81-84 iepd]
7 c2la7A_
90.2
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein yp_557733.1 from burkholderia xenovorans
8 c2ktsA_
82.4
34
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hslj;PDBTitle: nmr structure of the protein np_415897.1
9 c1uw7A_
29.8
19
PDB header: replicase proteinChain: A: PDB Molecule: nsp9;PDBTitle: nsp9 protein from sars-coronavirus.
10 d2bhua1
25.5
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes11 d1qz8a_
22.2
19
Fold: Replicase NSP9Superfamily: Replicase NSP9Family: Replicase NSP912 c3ff6D_
19.0
29
PDB header: ligaseChain: D: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: human acc2 ct domain with cp-640186
13 c1od4C_
18.4
29
PDB header: ligaseChain: C: PDB Molecule: acetyl-coenzyme a carboxylase;PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
14 d1gl0i_
18.4
38
Fold: PMP inhibitorsSuperfamily: PMP inhibitorsFamily: PMP inhibitors15 d1wo9a_
16.6
46
Fold: PMP inhibitorsSuperfamily: PMP inhibitorsFamily: PMP inhibitors16 d2qyqa1
16.6
20
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein17 c2gzqA_
16.4
22
PDB header: lipid binding proteinChain: A: PDB Molecule: phosphatidylethanolamine-binding protein;PDBTitle: phosphatidylethanolamine-binding protein from plasmodium vivax
18 c2f91B_
16.3
46
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: serine protease inhibitor i/ii;PDBTitle: 1.2a resolution structure of a crayfish trypsin complexed2 with a peptide inhibitor, sgti
19 d1uyra2
15.7
29
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain20 c1wkpA_
15.2
36
PDB header: signaling proteinChain: A: PDB Molecule: flowering locus t protein;PDBTitle: flowering locus t (ft) from arabidopsis thaliana
21 c3h0jA_
not modelled
13.9
29
PDB header: transferaseChain: A: PDB Molecule: acetyl-coa carboxylase;PDBTitle: crystal structure of the carboxyltransferase domain of2 acetyl-coenzyme a carboxylase in complex with compound 2
22 c3c5pF_
not modelled
13.7
17
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: protein bas0735 of unknown function;PDBTitle: crystal structure of bas0735, a protein of unknown function from2 bacillus anthracis str. sterne
23 c2jyzA_
not modelled
12.6
22
PDB header: unknown functionChain: A: PDB Molecule: cg7054-pa;PDBTitle: cg7054 solution structure
24 c2xu8B_
not modelled
12.1
30
PDB header: structural genomicsChain: B: PDB Molecule: pa1645;PDBTitle: structure of pa1645
25 d1igqa_
not modelled
11.7
28
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Transcriptional repressor protein KorB26 d1igub_
not modelled
11.6
28
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Transcriptional repressor protein KorB27 c2dreA_
not modelled
10.4
14
PDB header: plant proteinChain: A: PDB Molecule: water-soluble chlorophyll protein;PDBTitle: crystal structure of water-soluble chlorophyll protein from2 lepidium virginicum at 2.00 angstrom resolution
28 c1uytC_
not modelled
10.3
29
PDB header: transferaseChain: C: PDB Molecule: acetyl-coa carboxylase;PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
29 d1kn3a_
not modelled
10.2
22
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein30 d1qoua_
not modelled
10.0
36
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein31 d1khua_
not modelled
9.7
35
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain32 c2r77A_
not modelled
9.6
17
PDB header: lipid binding proteinChain: A: PDB Molecule: phosphatidylethanolamine-binding protein, putative;PDBTitle: crystal structure of phosphatidylethanolamine-binding protein,2 pfl0955c, from plasmodium falciparum
33 d1wpxb1
not modelled
9.6
26
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein34 d1dd1a_
not modelled
9.2
47
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain35 d1zxoa2
not modelled
9.1
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like36 d1a44a_
not modelled
9.1
22
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein37 c4a1bB_
not modelled
8.3
38
PDB header: ribosomeChain: B: PDB Molecule: rpl39;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 3.
38 c2l72A_
not modelled
8.0
33
PDB header: protein bindingChain: A: PDB Molecule: actin depolymerizing factor, putative;PDBTitle: solution structure and dynamics of adf from toxoplasma gondii (tgadf)
39 d1o5ua_
not modelled
7.6
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111240 d1ygsa_
not modelled
7.6
47
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain41 d1zbsa1
not modelled
7.5
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like42 d1xwva_
not modelled
7.3
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: ML domain43 d1u3em2
not modelled
7.1
43
Fold: DNA-binding domain of intron-encoded endonucleasesSuperfamily: DNA-binding domain of intron-encoded endonucleasesFamily: DNA-binding domain of intron-encoded endonucleases44 c3a9lB_
not modelled
6.9
33
PDB header: hydrolaseChain: B: PDB Molecule: poly-gamma-glutamate hydrolase;PDBTitle: structure of bacteriophage poly-gamma-glutamate hydrolase
45 d1khxa_
not modelled
6.9
41
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain46 c1khxA_
not modelled
6.9
41
PDB header: transcriptionChain: A: PDB Molecule: smad2;PDBTitle: crystal structure of a phosphorylated smad2
47 c2jv4A_
not modelled
6.5
22
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis/trans isomerase;PDBTitle: structure characterisation of pina ww domain and comparison2 with other group iv ww domains, pin1 and ess1
48 d1jjcb3
not modelled
6.3
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Myf domain49 d2jnaa1
not modelled
6.1
46
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like50 c3q1jA_
not modelled
6.0
21
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
51 c2zkr3_
not modelled
5.8
83
PDB header: ribosomal protein/rnaChain: 3: PDB Molecule: 60s ribosomal protein l39e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
52 c2x24B_
not modelled
5.3
24
PDB header: ligaseChain: B: PDB Molecule: acetyl-coa carboxylase;PDBTitle: bovine acc2 ct domain in complex with inhibitor