| 1 |
|
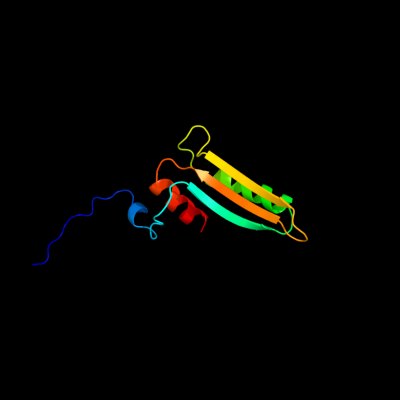
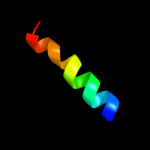
PDB 2k3i chain A
Region: 3 - 94
Aligned: 92
Modelled: 92
Confidence: 100.0%
Identity: 40%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yiis;
PDBTitle: solution nmr structure of protein yiis from shigella2 flexneri. northeast structural genomics consortium target3 sfr90
Phyre2
| 2 |
|
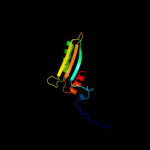
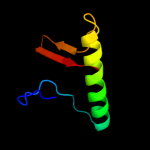
PDB 2jz5 chain A
Region: 14 - 94
Aligned: 79
Modelled: 81
Confidence: 100.0%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vpa0419;
PDBTitle: nmr solution structure of protein vpa0419 from vibrio2 parahaemolyticus. northeast structural genomics target3 vpr68
Phyre2
| 3 |
|
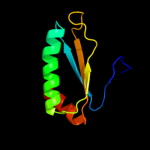
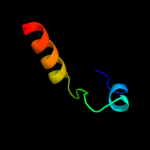
PDB 2khd chain A
Region: 1 - 94
Aligned: 91
Modelled: 94
Confidence: 100.0%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vc_a0919;
PDBTitle: solution nmr structure of vc_a0919 from vibrio cholerae.2 northeast structural genomics consortium target vcr52
Phyre2
| 4 |
|
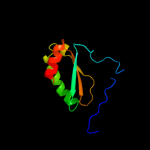
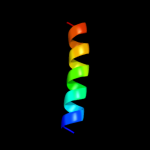
PDB 2o6n chain A
Region: 38 - 54
Aligned: 17
Modelled: 17
Confidence: 22.0%
Identity: 29%
PDB header:de novo protein
Chain: A: PDB Molecule:rh4b designed peptide;
PDBTitle: rh4b: designed right-handed coiled coil tetramer with all biological2 amino acids
Phyre2
| 5 |
|
PDB 1urz chain C
Region: 57 - 92
Aligned: 33
Modelled: 36
Confidence: 15.9%
Identity: 21%
PDB header:virus/viral protein
Chain: C: PDB Molecule:envelope protein;
PDBTitle: low ph induced, membrane fusion conformation of the2 envelope protein of tick-borne encephalitis virus
Phyre2
| 6 |
|
PDB 2yxy chain A
Region: 36 - 55
Aligned: 20
Modelled: 20
Confidence: 15.1%
Identity: 50%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical conserved protein, gk0453;
PDBTitle: crystarl structure of hypothetical conserved protein, gk0453
Phyre2
| 7 |
|
PDB 2k8e chain A
Region: 20 - 76
Aligned: 51
Modelled: 57
Confidence: 10.6%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0339 protein yegp;
PDBTitle: solution nmr structure of protein of unknown function yegp from e.2 coli. ontario center for structural proteomics target ec0640_1_1233 northeast structural genomics consortium target et102.
Phyre2
| 8 |
|
PDB 3i08 chain D
Region: 21 - 50
Aligned: 30
Modelled: 30
Confidence: 10.0%
Identity: 27%
PDB header:signaling protein
Chain: D: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: crystal structure of the s1-cleaved notch1 negative2 regulatory region (nrr)
Phyre2
| 9 |
|
PDB 3d3r chain A domain 1
Region: 36 - 55
Aligned: 20
Modelled: 20
Confidence: 10.0%
Identity: 30%
Fold: OB-fold
Superfamily: HupF/HypC-like
Family: HupF/HypC-like
Phyre2
| 10 |
|
PDB 1sf9 chain A
Region: 36 - 55
Aligned: 20
Modelled: 20
Confidence: 9.4%
Identity: 40%
Fold: SH3-like barrel
Superfamily: Hypothetical protein YfhH
Family: Hypothetical protein YfhH
Phyre2
| 11 |
|
PDB 3uaj chain A
Region: 18 - 92
Aligned: 38
Modelled: 38
Confidence: 7.5%
Identity: 8%
PDB header:viral protein/immune system
Chain: A: PDB Molecule:envelope protein;
PDBTitle: crystal structure of the envelope glycoprotein ectodomain from dengue2 virus serotype 4 in complex with the fab fragment of the chimpanzee3 monoclonal antibody 5h2
Phyre2
| 12 |
|
PDB 3d3r chain A
Region: 36 - 55
Aligned: 20
Modelled: 20
Confidence: 6.8%
Identity: 30%
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
Phyre2