| 1 |
|
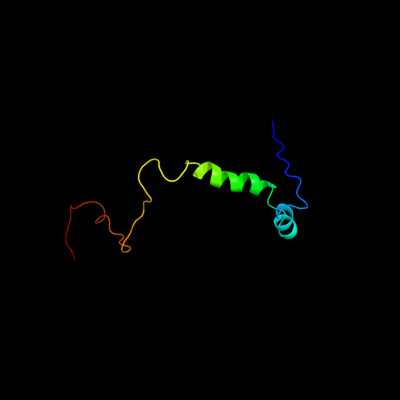
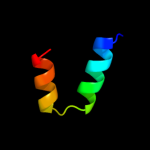
PDB 2adl chain B
Region: 1 - 72
Aligned: 72
Modelled: 72
Confidence: 100.0%
Identity: 96%
PDB header:dna binding protein
Chain: B: PDB Molecule:ccda;
PDBTitle: solution structure of the bacterial antitoxin ccda:2 implications for dna and toxin binding
Phyre2
| 2 |
|
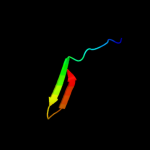
PDB 3g7z chain D
Region: 40 - 72
Aligned: 33
Modelled: 33
Confidence: 99.8%
Identity: 100%
PDB header:toxin/toxin repressor
Chain: D: PDB Molecule:protein ccda;
PDBTitle: ccdb dimer in complex with two c-terminal ccda domains
Phyre2
| 3 |
|
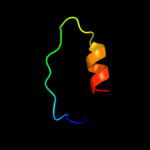
PDB 2xzm chain V
Region: 7 - 31
Aligned: 25
Modelled: 25
Confidence: 23.1%
Identity: 44%
PDB header:ribosome
Chain: V: PDB Molecule:rps17e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 4 |
|
PDB 2zou chain B
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 12.6%
Identity: 30%
PDB header:cell adhesion
Chain: B: PDB Molecule:spondin-1;
PDBTitle: crystal struture of human f-spondin reeler domain (fragment 2)
Phyre2
| 5 |
|
PDB 2db7 chain A domain 1
Region: 49 - 59
Aligned: 11
Modelled: 11
Confidence: 11.5%
Identity: 55%
Fold: Orange domain-like
Superfamily: Orange domain-like
Family: Hairy Orange domain
Phyre2
| 6 |
|
PDB 3coo chain B
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 10.6%
Identity: 30%
PDB header:cell adhesion
Chain: B: PDB Molecule:spondin-1;
PDBTitle: the crystal structure of reelin-n domain of f-spondin
Phyre2
| 7 |
|
PDB 1qk9 chain A
Region: 52 - 72
Aligned: 21
Modelled: 21
Confidence: 9.5%
Identity: 24%
Fold: DNA-binding domain
Superfamily: DNA-binding domain
Family: Methyl-CpG-binding domain, MBD
Phyre2
| 8 |
|
PDB 3d0w chain D
Region: 33 - 59
Aligned: 27
Modelled: 27
Confidence: 6.6%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:yflh protein;
PDBTitle: crystal structure of yflh protein from bacillus subtilis.2 northeast structural genomics consortium target sr326
Phyre2
| 9 |
|
PDB 2i3e chain A
Region: 18 - 37
Aligned: 20
Modelled: 20
Confidence: 5.7%
Identity: 30%
PDB header:hydrolase
Chain: A: PDB Molecule:g-rich;
PDBTitle: solution structure of catalytic domain of goldfish rich2 protein
Phyre2
| 10 |
|
PDB 1wz3 chain A domain 1
Region: 36 - 60
Aligned: 25
Modelled: 25
Confidence: 5.4%
Identity: 24%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Ubiquitin-like
Family: APG12-like
Phyre2