| 1 | d1jw2a_
|
|
 |
100.0 |
40 |
Fold:Open three-helical up-and-down bundle
Superfamily:Hemolysin expression modulating protein HHA
Family:Hemolysin expression modulating protein HHA |
| 2 | c2jqtA_
|
|
 |
99.9 |
100 |
PDB header:protein binding
Chain: A: PDB Molecule:h-ns/stpa-binding protein 2;
PDBTitle: structure of the bacterial replication origin-associated2 protein cnu
|
| 3 | c2jpnA_
|
|
 |
36.6 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent dna helicase uvsw;
PDBTitle: solution structure of t4 bacteriophage helicase uvsw.1
|
| 4 | c3jvoA_
|
|
 |
31.9 |
21 |
PDB header:viral protein
Chain: A: PDB Molecule:gp6;
PDBTitle: crystal structure of bacteriophage hk97 gp6
|
| 5 | d1lr1a_
|
|
 |
28.0 |
25 |
Fold:H-NS histone-like proteins
Superfamily:H-NS histone-like proteins
Family:H-NS histone-like proteins |
| 6 | c3ke4B_
|
|
 |
25.2 |
31 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical cytosolic protein;
PDBTitle: crystal structure of a pduo-type atp:cob(i)alamin adenosyltransferase2 from bacillus cereus
|
| 7 | c2idxA_
|
|
 |
23.6 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:cob(i)yrinic acid a,c-diamide
PDBTitle: structure of human atp:cobalamin adenosyltransferase bound2 to atp.
|
| 8 | d1trra_
|
|
 |
23.5 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Trp repressor, TrpR |
| 9 | d1ej2a_
|
|
 |
21.8 |
22 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 10 | d1jhga_
|
|
 |
12.8 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:TrpR-like
Family:Trp repressor, TrpR |
| 11 | c3cvfA_
|
|
 |
11.8 |
27 |
PDB header:signaling protein
Chain: A: PDB Molecule:homer protein homolog 3;
PDBTitle: crystal structure of the carboxy terminus of homer3
|
| 12 | c3d0jA_
|
|
 |
10.6 |
36 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ca_c3497;
PDBTitle: crystal structure of conserved protein of unknown function ca_c34972 from clostridium acetobutylicum atcc 824
|
| 13 | c2g2dA_
|
|
 |
10.3 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:atp:cobalamin adenosyltransferase;
PDBTitle: crystal structure of a putative pduo-type atp:cobalamin2 adenosyltransferase from mycobacterium tuberculosis
|
| 14 | c3cm8A_
|
|
 |
9.8 |
18 |
PDB header:rna binding protein/transferase
Chain: A: PDB Molecule:polymerase acidic protein;
PDBTitle: a rna polymerase subunit structure from virus
|
| 15 | d1t07a_
|
|
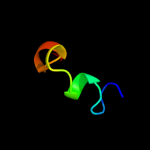 |
9.7 |
19 |
Fold:YggX-like
Superfamily:YggX-like
Family:YggX-like |
| 16 | d1xs8a_
|
|
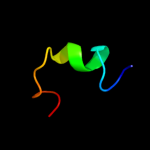 |
9.4 |
38 |
Fold:YggX-like
Superfamily:YggX-like
Family:YggX-like |
| 17 | c3i5qA_
|
|
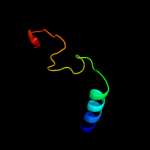 |
9.3 |
24 |
PDB header:protein transport
Chain: A: PDB Molecule:nucleoporin nup170;
PDBTitle: nup170(aa1253-1502) at 2.2 a, s.cerevisiae
|
| 18 | c2rbgB_
|
|
 |
8.9 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein st0493;
PDBTitle: crystal structure of hypothetical protein(st0493) from2 sulfolobus tokodaii
|
| 19 | c1nybA_
|
|
 |
8.5 |
42 |
PDB header:transcription/rna
Chain: A: PDB Molecule:probable regulatory protein n;
PDBTitle: solution structure of the bacteriophage phi21 n peptide-boxb2 rna complex
|
| 20 | d1dd4c_
|
|
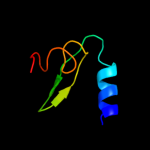 |
8.4 |
26 |
Fold:Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily:Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family:Ribosomal protein L7/12, oligomerisation (N-terminal) domain |
| 21 | d1od6a_ |
|
not modelled |
8.2 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 22 | c2nt8A_ |
|
not modelled |
7.8 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:cobalamin adenosyltransferase;
PDBTitle: atp bound at the active site of a pduo type atp:co(i)rrinoid2 adenosyltransferase from lactobacillus reuteri
|
| 23 | c2zhzC_ |
|
not modelled |
7.7 |
31 |
PDB header:transferase
Chain: C: PDB Molecule:atp:cob(i)alamin adenosyltransferase, putative;
PDBTitle: crystal structure of a pduo-type atp:cobalamin adenosyltransferase2 from burkholderia thailandensis
|
| 24 | c3nd5D_ |
|
not modelled |
7.5 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: crystal structure of phosphopantetheine adenylyltransferase (ppat)2 from enterococcus faecalis
|
| 25 | d1vlha_ |
|
not modelled |
7.5 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 26 | d1ni8a_ |
|
not modelled |
7.2 |
21 |
Fold:H-NS histone-like proteins
Superfamily:H-NS histone-like proteins
Family:H-NS histone-like proteins |
| 27 | d1uhra_ |
|
not modelled |
7.1 |
21 |
Fold:SWIB/MDM2 domain
Superfamily:SWIB/MDM2 domain
Family:SWIB/MDM2 domain |
| 28 | c2h29A_ |
|
not modelled |
7.0 |
60 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide
PDBTitle: crystal structure of nicotinic acid mononucleotide2 adenylyltransferase from staphylococcus aureus: product3 bound form 1
|
| 29 | c3frwF_ |
|
not modelled |
7.0 |
19 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative trp repressor protein;
PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
|
| 30 | d1f3ua_ |
|
not modelled |
6.7 |
40 |
Fold:triple barrel
Superfamily:Rap30/74 interaction domains
Family:Rap30/74 interaction domains |
| 31 | d1tfua_ |
|
not modelled |
6.4 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 32 | d1i27a_ |
|
not modelled |
6.1 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal domain of the rap74 subunit of TFIIF |
| 33 | d1v31a_ |
|
not modelled |
5.9 |
21 |
Fold:SWIB/MDM2 domain
Superfamily:SWIB/MDM2 domain
Family:SWIB/MDM2 domain |
| 34 | c1dd3D_ |
|
not modelled |
5.9 |
26 |
PDB header:ribosome
Chain: D: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
|
| 35 | c1dd3C_ |
|
not modelled |
5.9 |
26 |
PDB header:ribosome
Chain: C: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
|
| 36 | c3f3mA_ |
|
not modelled |
5.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphopantetheine adenylyltransferase;
PDBTitle: six crystal structures of two phosphopantetheine2 adenylyltransferases reveal an alternative ligand binding3 mode and an associated structural change
|
| 37 | d1o6ba_ |
|
not modelled |
5.7 |
18 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 38 | c2l3lA_ |
|
not modelled |
5.6 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:tubulin-specific chaperone c;
PDBTitle: the solution structure of the n-terminal domain of human tubulin2 binding cofactor c reveals a platform for the interaction with ab-3 tubulin
|
| 39 | c1zawV_ |
|
not modelled |
5.5 |
26 |
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
|
| 40 | c2kngA_ |
|
not modelled |
5.5 |
33 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein lsr2;
PDBTitle: solution structure of c-domain of lsr2
|
| 41 | c3korD_ |
|
not modelled |
5.4 |
18 |
PDB header:transcription
Chain: D: PDB Molecule:possible trp repressor;
PDBTitle: crystal structure of a putative trp repressor from staphylococcus2 aureus
|
| 42 | d1qjca_ |
|
not modelled |
5.3 |
11 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Adenylyltransferase |
| 43 | c1zawW_ |
|
not modelled |
5.3 |
26 |
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
|
| 44 | c1zawU_ |
|
not modelled |
5.3 |
26 |
PDB header:structural protein
Chain: U: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
|
| 45 | c1xhmB_ |
|
not modelled |
5.2 |
27 |
PDB header:signaling protein
Chain: B: PDB Molecule:guanine nucleotide-binding protein g(i)/g(s)
PDBTitle: the crystal structure of a biologically active peptide2 (sigk) bound to a g protein beta:gamma heterodimer
|
| 46 | c1zaxV_ |
|
not modelled |
5.1 |
26 |
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
|
| 47 | c1zavV_ |
|
not modelled |
5.1 |
26 |
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
|
| 48 | c1zaxY_ |
|
not modelled |
5.1 |
26 |
PDB header:structural protein
Chain: Y: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
|
| 49 | c1zavW_ |
|
not modelled |
5.1 |
26 |
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
|
| 50 | c1zaxW_ |
|
not modelled |
5.1 |
26 |
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
|
| 51 | c1zavX_ |
|
not modelled |
5.1 |
26 |
PDB header:structural protein
Chain: X: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
|





















































































