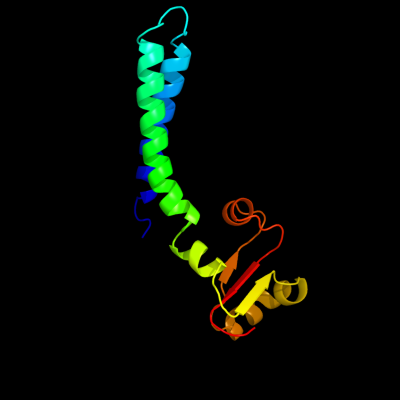
1 c2lf0A_
100.0
98
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yibl;PDBTitle: solution structure of sf3636, a two-domain unknown function protein2 from shigella flexneri 2a, determined by joint refinement of nmr,3 residual dipolar couplings and small-angle x-ray scatting, nesg4 target sfr339/ocsp target sf3636
2 c1gk7A_
91.4
36
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 1a fragment (1a)
3 c1t3jA_
78.5
33
PDB header: membrane proteinChain: A: PDB Molecule: mitofusin 1;PDBTitle: mitofusin domain hr2 v686m/i708m mutant
4 c2a3dA_
68.3
32
PDB header: three-helix bundleChain: A: PDB Molecule: protein (de novo three-helix bundle);PDBTitle: solution structure of a de novo designed single chain three-2 helix bundle (a3d)
5 c1u0iA_
65.6
67
PDB header: de novo proteinChain: A: PDB Molecule: iaal-e3;PDBTitle: iaal-e3/k3 heterodimer
6 c2c5iT_
60.3
14
PDB header: protein transportChain: T: PDB Molecule: t-snare affecting a late golgi compartmentPDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51 in distorted conformation
7 c1ce0B_
40.4
41
PDB header: hiv-1 envelope proteinChain: B: PDB Molecule: protein (leucine zipper model h38-p1);PDBTitle: trimerization specificity in hiv-1 gp41: analysis with a2 gcn4 leucine zipper model
8 c1g6uB_
30.8
26
PDB header: de novo proteinChain: B: PDB Molecule: domain swapped dimer;PDBTitle: crystal structure of a domain swapped dimer
9 c2xzrA_
29.2
27
PDB header: cell adhesionChain: A: PDB Molecule: immunoglobulin-binding protein eibd;PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
10 c1u2uA_
24.3
61
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: nmr solution structure of a designed heterodimeric leucine2 zipper
11 c1gp8A_
23.4
32
PDB header: viral proteinChain: A: PDB Molecule: protein (scaffolding protein);PDBTitle: nmr solution structure of the coat protein-binding domain2 of bacteriophage p22 scaffolding protein
12 c1ij2C_
23.0
45
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
13 c1ztaA_
21.7
45
PDB header: dna-binding motifChain: A: PDB Molecule: leucine zipper monomer;PDBTitle: the solution structure of a leucine-zipper motif peptide
14 c1ij2B_
21.1
45
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
15 c3u1aC_
20.7
23
PDB header: contractile proteinChain: C: PDB Molecule: smooth muscle tropomyosin alpha;PDBTitle: n-terminal 81-aa fragment of smooth muscle tropomyosin alpha
16 c1ic2B_
19.3
24
PDB header: contractile proteinChain: B: PDB Molecule: tropomyosin alpha chain, skeletal muscle;PDBTitle: deciphering the design of the tropomyosin molecule
17 c1j1dF_
19.2
24
PDB header: contractile proteinChain: F: PDB Molecule: troponin i;PDBTitle: crystal structure of the 46kda domain of human cardiac2 troponin in the ca2+ saturated form
18 c1rb6C_
18.7
45
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal form
19 c1rb1A_
18.7
45
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
20 c3k7zB_
18.7
45
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
21 c1rb1B_
not modelled
18.7
45
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
22 c1swiA_
not modelled
18.7
45
PDB header: leucine zipperChain: A: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with2 benzene
23 c3k7zA_
not modelled
18.7
45
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
24 c1ij3B_
not modelled
18.7
45
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
25 c1ij3C_
not modelled
18.7
45
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
26 c2ekkA_
not modelled
17.9
55
PDB header: protein bindingChain: A: PDB Molecule: uba domain from e3 ubiquitin-protein ligasePDBTitle: solution structure of ruh-074, a human uba domain
27 d1we3a2
not modelled
16.0
17
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain28 d1kida_
not modelled
16.0
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain29 c1wt6B_
not modelled
14.8
30
PDB header: transferaseChain: B: PDB Molecule: myotonin-protein kinase;PDBTitle: coiled-coil domain of dmpk
30 d1sjpa2
not modelled
14.5
17
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain31 c3he4A_
not modelled
14.5
59
PDB header: de novo proteinChain: A: PDB Molecule: synzip6;PDBTitle: heterospecific coiled-coil pair synzip5:synzip6
32 c2dq3A_
not modelled
13.9
23
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of aq_298
33 c1yv0I_
not modelled
13.8
22
PDB header: contractile proteinChain: I: PDB Molecule: troponin i, fast skeletal muscle;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-2 free state
34 c3m6cA_
not modelled
13.8
17
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 1;PDBTitle: crystal structure of mycobacterium tuberculosis groel1 apical domain
35 c2o7hF_
not modelled
13.5
41
PDB header: transcriptionChain: F: PDB Molecule: general control protein gcn4;PDBTitle: crystal structure of trimeric coiled coil gcn4 leucine zipper
36 d1dl5a2
not modelled
13.4
35
Fold: Protein-L-isoaspartyl O-methyltransferase, C-terminal domainSuperfamily: Protein-L-isoaspartyl O-methyltransferase, C-terminal domainFamily: Protein-L-isoaspartyl O-methyltransferase, C-terminal domain37 d1rq2a1
not modelled
13.2
32
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain38 c3ni0A_
not modelled
12.8
32
PDB header: immune systemChain: A: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: crystal structure of mouse bst-2/tetherin ectodomain
39 d2c0sa1
not modelled
12.5
18
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like40 c3nmdA_
not modelled
12.3
42
PDB header: transferaseChain: A: PDB Molecule: cgmp dependent protein kinase;PDBTitle: crystal structure of the leucine zipper domain of cgmp dependent2 protein kinase i beta
41 c2w9kA_
not modelled
11.4
25
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crithidia fasciculata cytochrome c
42 c2etnA_
not modelled
11.3
21
PDB header: transcriptionChain: A: PDB Molecule: anti-cleavage anti-grea transcription factorPDBTitle: crystal structure of thermus aquaticus gfh1
43 c3oa7A_
not modelled
10.9
16
PDB header: structural proteinChain: A: PDB Molecule: head morphogenesis protein, chaotic nuclear migrationPDBTitle: structure of the c-terminal domain of cnm67, a core component of the2 spindle pole body of saccharomyces cerevisiae
44 c4a19Q_
not modelled
10.7
44
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l36;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
45 c1ytzI_
not modelled
10.5
22
PDB header: contractile proteinChain: I: PDB Molecule: troponin i;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-2 activated state
46 c1gk4A_
not modelled
10.4
20
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment (cys2)
47 d2nzca1
not modelled
10.3
11
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: TM1266-like48 c3lpeF_
not modelled
10.3
17
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
49 d2g3qa1
not modelled
9.7
27
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain50 c3fgaD_
not modelled
9.2
20
PDB header: hydrolase/hydrolase inhibitorChain: D: PDB Molecule: shugoshin-like 1;PDBTitle: structural basis of pp2a and sgo interaction
51 c2kswA_
not modelled
9.1
50
PDB header: hydrolase inhibitorChain: A: PDB Molecule: oryctin;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for oryctin
52 d1ivsa1
not modelled
9.0
13
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain53 d5csma_
not modelled
8.8
17
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Allosteric chorismate mutase54 c3izck_
not modelled
8.7
33
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein rpl16 (l13p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
55 c2rklB_
not modelled
8.6
24
PDB header: lipid transportChain: B: PDB Molecule: vacuolar protein sorting-associated protein vta1;PDBTitle: crystal structure of s.cerevisiae vta1 c-terminal domain
56 d1uklc_
not modelled
8.6
9
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain57 c2l5gB_
not modelled
8.5
24
PDB header: transcription regulatorChain: B: PDB Molecule: putative uncharacterized protein ncor2;PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
58 d1gqea_
not modelled
8.1
15
Fold: Release factorSuperfamily: Release factorFamily: Release factor59 c1gcmA_
not modelled
7.9
38
PDB header: transcription regulationChain: A: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
60 c2zkrv_
not modelled
7.9
17
PDB header: ribosomal protein/rnaChain: V: PDB Molecule: rna expansion segment es9 part2;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
61 c1r48A_
not modelled
7.6
37
PDB header: transport proteinChain: A: PDB Molecule: proline/betaine transporter;PDBTitle: solution structure of the c-terminal cytoplasmic domain2 residues 468-497 of escherichia coli protein prop
62 c3a5tB_
not modelled
7.5
26
PDB header: transcription regulator/dnaChain: B: PDB Molecule: transcription factor mafg;PDBTitle: crystal structure of mafg-dna complex
63 c2wukD_
not modelled
7.5
29
PDB header: cell cycleChain: D: PDB Molecule: septum site-determining protein diviva;PDBTitle: diviva n-terminal domain, f17a mutant
64 c1kddC_
not modelled
7.5
56
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
65 c1kddF_
not modelled
7.3
56
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
66 c1kddA_
not modelled
7.3
56
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
67 c3ol1A_
not modelled
7.2
22
PDB header: structural proteinChain: A: PDB Molecule: vimentin;PDBTitle: crystal structure of vimentin (fragment 144-251) from homo sapiens,2 northeast structural genomics consortium target hr4796b
68 d1oqya1
not modelled
7.2
16
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain69 c1gcmB_
not modelled
7.1
38
PDB header: transcription regulationChain: B: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
70 c3tqmD_
not modelled
7.1
18
PDB header: protein bindingChain: D: PDB Molecule: ribosome-associated factor y;PDBTitle: structure of an ribosomal subunit interface protein from coxiella2 burnetii
71 c2hx6A_
not modelled
7.0
13
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
72 c2a45J_
not modelled
6.9
7
PDB header: hydrolase/hydrolase inhibitorChain: J: PDB Molecule: fibrinogen alpha chain;PDBTitle: crystal structure of the complex between thrombin and the central "e"2 region of fibrin
73 c2z9fC_
not modelled
6.8
17
PDB header: biosynthetic proteinChain: C: PDB Molecule: cellulose synthase operon protein d;PDBTitle: crystal structure of axcesd protein from acetobacter xylinum
74 c1gcmC_
not modelled
6.8
38
PDB header: transcription regulationChain: C: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
75 c1sryB_
not modelled
6.7
12
PDB header: ligase(synthetase)Chain: B: PDB Molecule: seryl-trna synthetase;PDBTitle: refined crystal structure of the seryl-trna synthetase from2 thermus thermophilus at 2.5 angstroms resolution
76 c3lt7D_
not modelled
6.5
29
PDB header: cell adhesionChain: D: PDB Molecule: adhesin yada;PDBTitle: a transition from strong right-handed to canonical left-handed2 supercoiling in a conserved coiled coil segment of trimeric3 autotransporter adhesins - the m3 mutant structure
77 d1gpja1
not modelled
6.4
20
Fold: Glutamyl tRNA-reductase dimerization domainSuperfamily: Glutamyl tRNA-reductase dimerization domainFamily: Glutamyl tRNA-reductase dimerization domain78 c2xu6B_
not modelled
6.3
22
PDB header: protein bindingChain: B: PDB Molecule: mdv1 coiled coil;PDBTitle: mdv1 coiled coil domain
79 c3h5fC_
not modelled
6.3
29
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l16l-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
80 c3h5gC_
not modelled
6.3
29
PDB header: de novo proteinChain: C: PDB Molecule: coil ser l16d-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
81 c3h5fA_
not modelled
6.3
29
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l16l-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
82 c3h5gA_
not modelled
6.3
29
PDB header: de novo proteinChain: A: PDB Molecule: coil ser l16d-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
83 c3h5fB_
not modelled
6.3
29
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l16l-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
84 c3h5gB_
not modelled
6.3
29
PDB header: de novo proteinChain: B: PDB Molecule: coil ser l16d-pen;PDBTitle: switching the chirality of the metal environment alters the2 coordination mode in designed peptides.
85 c2e43A_
not modelled
6.2
26
PDB header: transcription/dnaChain: A: PDB Molecule: ccaat/enhancer-binding protein beta;PDBTitle: crystal structure of c/ebpbeta bzip homodimer k269a mutant2 bound to a high affinity dna fragment
86 d1dp7p_
not modelled
6.2
55
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: P4 origin-binding domain-like87 c2ergA_
not modelled
6.1
22
PDB header: transcription activator/dnaChain: A: PDB Molecule: regulatory protein leu3;PDBTitle: crystal structure of leu3 dna-binding domain with a single2 h50c mutation complexed with a 15mer dna duplex
88 c2wt7B_
not modelled
6.1
19
PDB header: transcriptionChain: B: PDB Molecule: transcription factor mafb;PDBTitle: crystal structure of the bzip heterodimeric complex2 mafb:cfos bound to dna
89 c1grjA_
not modelled
5.9
7
PDB header: transcription regulationChain: A: PDB Molecule: grea protein;PDBTitle: grea transcript cleavage factor from escherichia coli
90 d1mska_
not modelled
5.9
45
Fold: Methionine synthase activation domain-likeSuperfamily: Methionine synthase activation domain-likeFamily: Methionine synthase SAM-binding domain91 c1ij2A_
not modelled
5.6
48
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
92 c1ofuB_
not modelled
5.5
32
PDB header: bacterial cell division inhibitorChain: B: PDB Molecule: cell division protein ftsz;PDBTitle: crystal structure of sula:ftsz from pseudomonas aeruginosa
93 c2vawA_
not modelled
5.4
32
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: ftsz pseudomonas aeruginosa gdp
94 c1u7mB_
not modelled
5.3
40
PDB header: de novo proteinChain: B: PDB Molecule: four-helix bundle model;PDBTitle: solution structure of a diiron protein model: due ferri(ii)2 turn mutant
95 c3he5A_
not modelled
5.3
32
PDB header: de novo proteinChain: A: PDB Molecule: synzip1;PDBTitle: heterospecific coiled-coil pair synzip2:synzip1