| 1 |
|
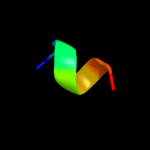
PDB 2nr1 chain A
Region: 24 - 28
Aligned: 5
Modelled: 5
Confidence: 18.1%
Identity: 80%
PDB header:receptor
Chain: A: PDB Molecule:nr1 m2;
PDBTitle: transmembrane segment 2 of nmda receptor nr1, nmr, 102 structures
Phyre2
| 2 |
|
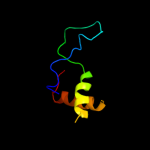
PDB 1ffg chain B
Region: 126 - 132
Aligned: 7
Modelled: 7
Confidence: 17.9%
Identity: 57%
Fold: Ferredoxin-like
Superfamily: CheY-binding domain of CheA
Family: CheY-binding domain of CheA
Phyre2
| 3 |
|
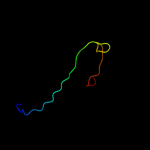
PDB 1a0o chain H
Region: 126 - 132
Aligned: 7
Modelled: 7
Confidence: 17.7%
Identity: 57%
PDB header:chemotaxis
Chain: H: PDB Molecule:chea;
PDBTitle: chey-binding domain of chea in complex with chey
Phyre2
| 4 |
|
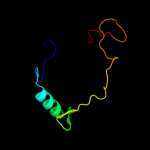
PDB 2ysr chain A
Region: 78 - 138
Aligned: 55
Modelled: 61
Confidence: 13.4%
Identity: 20%
PDB header:signaling protein
Chain: A: PDB Molecule:dep domain-containing protein 1;
PDBTitle: solution structure of the dep domain from human dep domain-2 containing protein 1
Phyre2
| 5 |
|
PDB 2ld7 chain A
Region: 71 - 90
Aligned: 19
Modelled: 20
Confidence: 11.6%
Identity: 47%
PDB header:transcription
Chain: A: PDB Molecule:histone deacetylase complex subunit sap30;
PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
Phyre2
| 6 |
|
PDB 3mqp chain B
Region: 72 - 82
Aligned: 11
Modelled: 11
Confidence: 11.5%
Identity: 73%
PDB header:apoptosis
Chain: B: PDB Molecule:phorbol-12-myristate-13-acetate-induced protein 1;
PDBTitle: crystal structure of human bfl-1 in complex with noxa bh3 peptide,2 northeast structural genomics consortium target hr2930
Phyre2
| 7 |
|
PDB 3kzw chain D
Region: 118 - 153
Aligned: 36
Modelled: 36
Confidence: 8.1%
Identity: 22%
PDB header:hydrolase
Chain: D: PDB Molecule:cytosol aminopeptidase;
PDBTitle: crystal structure of cytosol aminopeptidase from staphylococcus aureus2 col
Phyre2
| 8 |
|
PDB 2eqo chain A
Region: 30 - 37
Aligned: 8
Modelled: 8
Confidence: 8.0%
Identity: 63%
PDB header:transcription
Chain: A: PDB Molecule:tnf receptor-associated factor 3-interacting
PDBTitle: solution structure of the stn_traf3ip1_nd domain of2 interleukin 13 receptor alpha 1-binding protein-1 [homo3 sapiens]
Phyre2
| 9 |
|
PDB 1sg1 chain X domain 2
Region: 14 - 21
Aligned: 8
Modelled: 8
Confidence: 8.0%
Identity: 63%
Fold: TNF receptor-like
Superfamily: TNF receptor-like
Family: TNF receptor-like
Phyre2
| 10 |
|
PDB 1j3b chain A domain 2
Region: 54 - 124
Aligned: 71
Modelled: 71
Confidence: 7.0%
Identity: 21%
Fold: PEP carboxykinase N-terminal domain
Superfamily: PEP carboxykinase N-terminal domain
Family: PEP carboxykinase N-terminal domain
Phyre2
| 11 |
|
PDB 3im0 chain A
Region: 49 - 69
Aligned: 21
Modelled: 21
Confidence: 6.3%
Identity: 19%
PDB header:lyase
Chain: A: PDB Molecule:val-1;
PDBTitle: crystal structure of chlorella virus val-1 soaked in 200mm d-2 glucuronic acid, 10% peg-3350, and 200mm glycine-naoh (ph 10.0)
Phyre2
| 12 |
|
PDB 2iif chain A
Region: 94 - 108
Aligned: 15
Modelled: 15
Confidence: 6.0%
Identity: 40%
PDB header:recombination/dna
Chain: A: PDB Molecule:integration host factor;
PDBTitle: single chain integration host factor mutant protein (scihf2-2 k45ae) in complex with dna
Phyre2
| 13 |
|
PDB 1w4m chain A
Region: 107 - 138
Aligned: 32
Modelled: 32
Confidence: 5.8%
Identity: 22%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: DEP domain
Phyre2
| 14 |
|
PDB 2cso chain A domain 1
Region: 107 - 138
Aligned: 32
Modelled: 32
Confidence: 5.7%
Identity: 22%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: DEP domain
Phyre2
| 15 |
|
PDB 3hlu chain A
Region: 108 - 128
Aligned: 21
Modelled: 21
Confidence: 5.6%
Identity: 33%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein duf2179;
PDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 duf2179 from eubacterium ventriosum
Phyre2
| 16 |
|
PDB 1lan chain A
Region: 118 - 153
Aligned: 36
Modelled: 36
Confidence: 5.6%
Identity: 19%
PDB header:hydrolase (alpha-aminoacylpeptide)
Chain: A: PDB Molecule:leucine aminopeptidase;
PDBTitle: leucine aminopeptidase complex with l-leucinal
Phyre2
| 17 |
|
PDB 1kjs chain A
Region: 130 - 138
Aligned: 9
Modelled: 9
Confidence: 5.6%
Identity: 33%
Fold: Anaphylotoxins (complement system)
Superfamily: Anaphylotoxins (complement system)
Family: Anaphylotoxins (complement system)
Phyre2
| 18 |
|
PDB 1gxi chain E
Region: 133 - 147
Aligned: 15
Modelled: 15
Confidence: 5.5%
Identity: 60%
Fold: SH3-like barrel
Superfamily: Electron transport accessory proteins
Family: Photosystem I accessory protein E (PsaE)
Phyre2
| 19 |
|
PDB 1pse chain A
Region: 133 - 147
Aligned: 15
Modelled: 15
Confidence: 5.4%
Identity: 60%
Fold: SH3-like barrel
Superfamily: Electron transport accessory proteins
Family: Photosystem I accessory protein E (PsaE)
Phyre2
| 20 |
|
PDB 1k8r chain B
Region: 18 - 28
Aligned: 11
Modelled: 11
Confidence: 5.4%
Identity: 64%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Ubiquitin-like
Family: Ras-binding domain, RBD
Phyre2
| 21 |
|
| 22 |
|