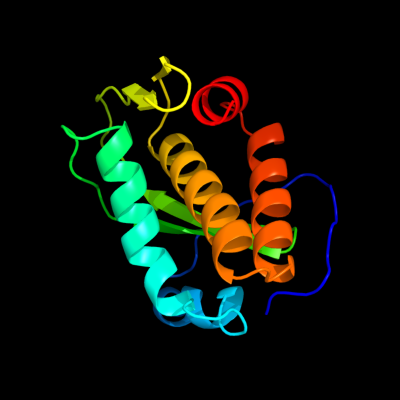
1 c2kmgA_
100.0
38
PDB header: gene regulationChain: A: PDB Molecule: klca;PDBTitle: the structure of the klca and ardb proteins show a novel2 fold and antirestriction activity against type i dna3 restriction systems in vivo but not in vitro
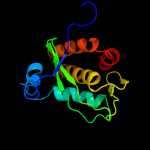
2 c2wj9A_
100.0
73
PDB header: hydrolase inhibitorChain: A: PDB Molecule: intergenic-region protein;PDBTitle: ardb
3 d2v6ai1
42.9
25
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit4 d1gk8i_
38.2
25
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit5 d8ruci_
33.0
33
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit6 d1wdds_
31.7
33
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit7 d1uzdc1
31.4
33
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit8 d1ej7s_
30.1
33
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit9 d2tssa2
25.3
29
Fold: beta-Grasp (ubiquitin-like)Superfamily: Superantigen toxins, C-terminal domainFamily: Superantigen toxins, C-terminal domain10 d1uzhc1
23.9
17
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit11 d1x3zb1
23.4
25
Fold: XPC-binding domainSuperfamily: XPC-binding domainFamily: XPC-binding domain12 d1ir1s_
23.2
33
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit13 c2qsgX_
23.0
25
PDB header: dna binding protein/dnaChain: X: PDB Molecule: uv excision repair protein rad23;PDBTitle: crystal structure of rad4-rad23 bound to a uv-damaged dna
14 d1m15a1
22.1
36
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain15 d1qh4a1
19.5
21
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain16 c2otnB_
18.9
15
PDB header: isomeraseChain: B: PDB Molecule: diaminopimelate epimerase;PDBTitle: crystal structure of the catalytically active form of diaminopimelate2 epimerase from bacillus anthracis
17 d1vrpa1
18.6
21
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain18 d1ks8a_
18.6
7
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Cellulases catalytic domain19 d1u6ra1
18.4
21
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain20 d1i0ea1
17.6
21
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain21 d1crka1
not modelled
17.6
14
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain22 d1g0wa1
not modelled
17.5
21
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain23 d1rbli_
not modelled
17.4
17
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit24 d1qk1a1
not modelled
17.0
21
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain25 d1svdm1
not modelled
16.3
14
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit26 d1bxni_
not modelled
16.2
17
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit27 d1bwvs_
not modelled
15.9
17
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit28 d1a7ha_
not modelled
13.3
19
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins29 c3lo4B_
not modelled
13.1
44
PDB header: antimicrobial proteinChain: B: PDB Molecule: neutrophil defensin 1;PDBTitle: crystal structure of human alpha-defensin 1 (r24a mutant)
30 c3lo4A_
not modelled
13.1
44
PDB header: antimicrobial proteinChain: A: PDB Molecule: neutrophil defensin 1;PDBTitle: crystal structure of human alpha-defensin 1 (r24a mutant)
31 c3cinA_
not modelled
12.2
23
PDB header: isomeraseChain: A: PDB Molecule: myo-inositol-1-phosphate synthase-related protein;PDBTitle: crystal structure of a myo-inositol-1-phosphate synthase-related2 protein (tm_1419) from thermotoga maritima msb8 at 1.70 a resolution
32 d1elpa2
not modelled
11.5
22
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins33 c3aq3A_
not modelled
11.4
39
PDB header: toxinChain: A: PDB Molecule: 6b protein;PDBTitle: molecular insights into plant cell proliferation disturbance by2 agrobacterium protein 6b
34 d1zira2
not modelled
10.7
19
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins35 d1enfa2
not modelled
10.2
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: Superantigen toxins, C-terminal domainFamily: Superantigen toxins, C-terminal domain36 d1kfia3
not modelled
9.8
18
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains37 d1ia9a_
not modelled
9.5
18
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: MHCK/EF2 kinase38 d1bi5a2
not modelled
9.5
15
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like39 d1amma2
not modelled
9.4
16
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins40 c1xxgA_
not modelled
9.1
29
PDB header: immune systemChain: A: PDB Molecule: enterotoxin;PDBTitle: crystal structure of staphylococcal enterotoxin g
41 d1y0ua_
not modelled
8.1
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators42 c1ck1A_
not modelled
8.1
41
PDB header: toxinChain: A: PDB Molecule: protein (enterotoxin type c-3);PDBTitle: structure of staphylococcal enterotoxin c3
43 c2xjqA_
not modelled
7.6
16
PDB header: cell adhesionChain: A: PDB Molecule: flocculation protein flo5;PDBTitle: x-ray structure of the n-terminal domain of the flocculin2 flo5 from saccharomyces cerevisiae
44 d2hg7a1
not modelled
7.5
30
Fold: gpW/XkdW-likeSuperfamily: XkdW-likeFamily: XkdW-like45 c2hg7A_
not modelled
7.5
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage-like element pbsx protein xkdw;PDBTitle: solution nmr structure of phage-like element pbsx protein2 xkdw, northeast structural genomics consortium target sr355
46 c1d6eC_
not modelled
7.4
31
PDB header: immune system/peptide inhibitorChain: C: PDB Molecule: enterotoxin type b;PDBTitle: crystal structure of hla-dr4 complex with peptidomimetic and seb
47 d1uaya_
not modelled
7.4
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases48 d3pmga3
not modelled
7.3
21
Fold: Phosphoglucomutase, first 3 domainsSuperfamily: Phosphoglucomutase, first 3 domainsFamily: Phosphoglucomutase, first 3 domains49 d1j83a_
not modelled
7.2
19
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 17 carbohydrate binding module, CBM1750 d1okia2
not modelled
6.8
13
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins51 d1ytqa2
not modelled
6.5
8
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins52 d1ueba2
not modelled
6.2
7
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like53 c2k1wA_
not modelled
6.1
23
PDB header: metal binding proteinChain: A: PDB Molecule: beta/gama crystallin family protein;PDBTitle: nmr solution structure of m-crystallin in calcium loaded2 form(holo).
54 c2j4xA_
not modelled
6.1
12
PDB header: signaling proteinChain: A: PDB Molecule: mitogen;PDBTitle: streptococcus dysgalactiae-derived mitogen (sdm)
55 d1h4ax1
not modelled
6.1
13
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins56 d2z8la2
not modelled
6.1
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: Superantigen toxins, C-terminal domainFamily: Superantigen toxins, C-terminal domain57 c1et9A_
not modelled
6.1
24
PDB header: immune systemChain: A: PDB Molecule: superantigen spe-h;PDBTitle: crystal structure of the superantigen spe-h from2 streptococcus pyogenes
58 d1amma1
not modelled
6.0
13
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Crystallins/Ca-binding development proteins59 c1bxtA_
not modelled
5.8
35
PDB header: immune systemChain: A: PDB Molecule: protein (streptococcal superantigen);PDBTitle: streptococcal superantigen (ssa) from streptococcus pyogenes
60 d1x4ka2
not modelled
5.7
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain61 c1js4B_
not modelled
5.5
8
PDB header: glycosyl hydrolaseChain: B: PDB Molecule: endo/exocellulase e4;PDBTitle: endo/exocellulase:cellobiose from thermomonospora
62 d3buxb3
not modelled
5.2
38
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain