| 1 |
|
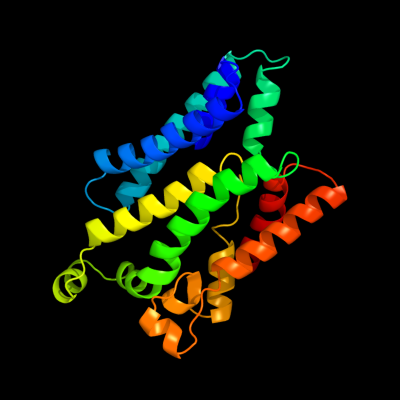
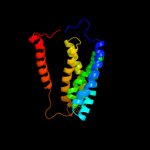
PDB 2nq2 chain A
Region: 90 - 320
Aligned: 218
Modelled: 219
Confidence: 97.9%
Identity: 11%
PDB header:metal transport
Chain: A: PDB Molecule:hypothetical abc transporter permease protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
Phyre2
| 2 |
|
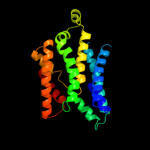
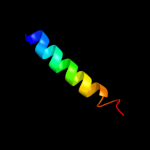
PDB 1l7v chain A
Region: 15 - 322
Aligned: 291
Modelled: 296
Confidence: 96.6%
Identity: 10%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 3 |
|
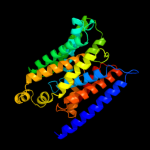
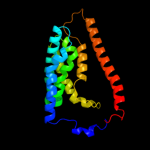
PDB 2b2h chain A
Region: 8 - 223
Aligned: 207
Modelled: 216
Confidence: 76.9%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 4 |
|
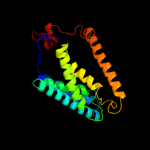
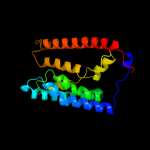
PDB 1u7g chain A
Region: 8 - 203
Aligned: 194
Modelled: 196
Confidence: 56.3%
Identity: 15%
Fold: Ammonium transporter
Superfamily: Ammonium transporter
Family: Ammonium transporter
Phyre2
| 5 |
|
PDB 2e74 chain G domain 1
Region: 229 - 252
Aligned: 24
Modelled: 24
Confidence: 21.0%
Identity: 25%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 6 |
|
PDB 1pgl chain 2 domain 2
Region: 1 - 16
Aligned: 16
Modelled: 16
Confidence: 18.4%
Identity: 13%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Comoviridae-like VP
Phyre2
| 7 |
|
PDB 1ny7 chain 2 domain 2
Region: 1 - 16
Aligned: 16
Modelled: 16
Confidence: 18.4%
Identity: 13%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Comoviridae-like VP
Phyre2
| 8 |
|
PDB 3hd6 chain A
Region: 8 - 201
Aligned: 189
Modelled: 194
Confidence: 16.5%
Identity: 13%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 9 |
|
PDB 3b9y chain A
Region: 7 - 202
Aligned: 184
Modelled: 196
Confidence: 14.5%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 10 |
|
PDB 3hd7 chain A
Region: 11 - 49
Aligned: 39
Modelled: 39
Confidence: 12.2%
Identity: 23%
PDB header:exocytosis
Chain: A: PDB Molecule:vesicle-associated membrane protein 2;
PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
Phyre2
| 11 |
|
PDB 3u5e chain L
Region: 202 - 221
Aligned: 20
Modelled: 20
Confidence: 9.8%
Identity: 30%
PDB header:ribosome
Chain: L: PDB Molecule:60s ribosomal protein l13-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
Phyre2
| 12 |
|
PDB 4a18 chain U
Region: 202 - 221
Aligned: 20
Modelled: 20
Confidence: 9.3%
Identity: 30%
PDB header:ribosome
Chain: U: PDB Molecule:rpl13;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
Phyre2
| 13 |
|
PDB 3pxp chain A
Region: 194 - 226
Aligned: 33
Modelled: 32
Confidence: 8.7%
Identity: 15%
PDB header:transcription regulator
Chain: A: PDB Molecule:helix-turn-helix domain protein;
PDBTitle: crystal structure of a pas and dna binding domain containing protein2 (caur_2278) from chloroflexus aurantiacus j-10-fl at 2.30 a3 resolution
Phyre2
| 14 |
|
PDB 1vf5 chain G
Region: 229 - 248
Aligned: 20
Modelled: 20
Confidence: 6.6%
Identity: 25%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 15 |
|
PDB 1vf5 chain G
Region: 229 - 248
Aligned: 20
Modelled: 20
Confidence: 6.6%
Identity: 25%
PDB header:photosynthesis
Chain: G: PDB Molecule:protein pet g;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
Phyre2
| 16 |
|
PDB 1y6u chain A
Region: 210 - 227
Aligned: 18
Modelled: 18
Confidence: 6.5%
Identity: 11%
PDB header:dna binding protein
Chain: A: PDB Molecule:excisionase from transposon tn916;
PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
Phyre2
| 17 |
|
PDB 1jb0 chain D
Region: 196 - 215
Aligned: 20
Modelled: 20
Confidence: 6.3%
Identity: 20%
Fold: Photosystem I subunit PsaD
Superfamily: Photosystem I subunit PsaD
Family: Photosystem I subunit PsaD
Phyre2
| 18 |
|
PDB 3e0d chain A
Region: 78 - 110
Aligned: 33
Modelled: 33
Confidence: 5.3%
Identity: 27%
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase iii subunit alpha;
PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
Phyre2