| 1 |
|
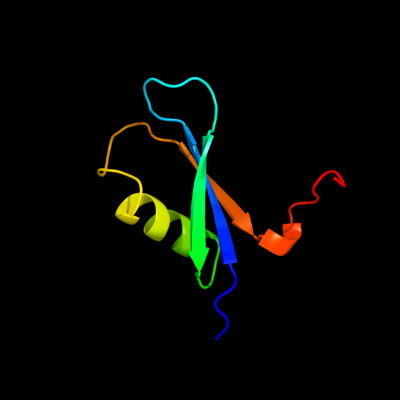
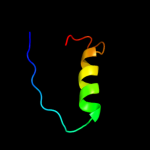
PDB 3egr chain B
Region: 4 - 65
Aligned: 62
Modelled: 62
Confidence: 100.0%
Identity: 65%
PDB header:oxidoreductase
Chain: B: PDB Molecule:phenylacetate-coa oxygenase subunit paab;
PDBTitle: crystal structure of a phenylacetate-coa oxygenase subunit paab2 (reut_a2307) from ralstonia eutropha jmp134 at 2.65 a resolution
Phyre2
| 2 |
|
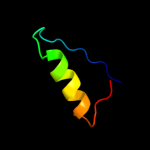
PDB 1na6 chain A domain 1
Region: 49 - 59
Aligned: 11
Modelled: 11
Confidence: 14.2%
Identity: 27%
Fold: DNA-binding pseudobarrel domain
Superfamily: DNA-binding pseudobarrel domain
Family: Type II restriction endonuclease effector domain
Phyre2
| 3 |
|
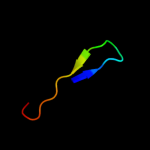
PDB 2fgc chain A domain 2
Region: 22 - 52
Aligned: 29
Modelled: 31
Confidence: 12.5%
Identity: 21%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: IlvH-like
Phyre2
| 4 |
|
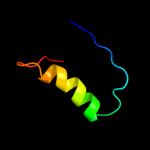
PDB 2f1f chain A domain 1
Region: 22 - 52
Aligned: 29
Modelled: 31
Confidence: 10.2%
Identity: 28%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: IlvH-like
Phyre2
| 5 |
|
PDB 2j6a chain A
Region: 83 - 95
Aligned: 13
Modelled: 13
Confidence: 9.7%
Identity: 31%
PDB header:transferase
Chain: A: PDB Molecule:protein trm112;
PDBTitle: crystal structure of s. cerevisiae ynr046w, a zinc finger2 protein from the erf1 methyltransferase complex.
Phyre2
| 6 |
|
PDB 2pc6 chain A domain 2
Region: 21 - 52
Aligned: 30
Modelled: 31
Confidence: 9.5%
Identity: 33%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: IlvH-like
Phyre2
| 7 |
|
PDB 1a0i chain A domain 1
Region: 68 - 85
Aligned: 18
Modelled: 18
Confidence: 7.9%
Identity: 22%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: DNA ligase/mRNA capping enzyme postcatalytic domain
Phyre2
| 8 |
|
PDB 2f1f chain A
Region: 22 - 52
Aligned: 29
Modelled: 31
Confidence: 6.8%
Identity: 28%
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
Phyre2
| 9 |
|
PDB 1wv8 chain A domain 1
Region: 44 - 60
Aligned: 16
Modelled: 17
Confidence: 6.5%
Identity: 19%
Fold: TTHA1013/TTHA0281-like
Superfamily: TTHA1013/TTHA0281-like
Family: TTHA1013-like
Phyre2
| 10 |
|
PDB 2pc6 chain C
Region: 22 - 52
Aligned: 29
Modelled: 31
Confidence: 6.2%
Identity: 34%
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
Phyre2
| 11 |
|
PDB 2gu3 chain A domain 1
Region: 7 - 18
Aligned: 12
Modelled: 12
Confidence: 6.0%
Identity: 42%
Fold: Cystatin-like
Superfamily: Cystatin/monellin
Family: PepSY-like
Phyre2