| 1 |
|
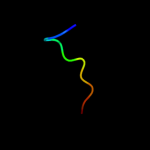
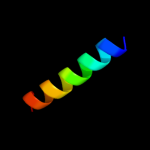
PDB 2rfp chain A
Region: 22 - 31
Aligned: 10
Modelled: 9
Confidence: 30.4%
Identity: 30%
PDB header:hydrolase
Chain: A: PDB Molecule:putative ntp pyrophosphohydrolase;
PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
Phyre2
| 2 |
|
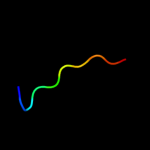
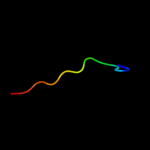
PDB 1gpi chain A
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 19.9%
Identity: 60%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Glycosyl hydrolase family 7 catalytic core
Phyre2
| 3 |
|
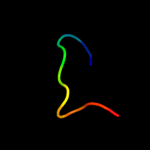
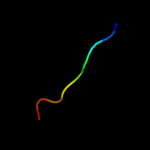
PDB 2v3i chain A domain 1
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 19.3%
Identity: 70%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Glycosyl hydrolase family 7 catalytic core
Phyre2
| 4 |
|
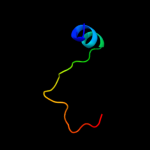
PDB 1q9h chain A
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 19.0%
Identity: 70%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Glycosyl hydrolase family 7 catalytic core
Phyre2
| 5 |
|
PDB 2rfy chain B
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 17.8%
Identity: 50%
PDB header:hydrolase
Chain: B: PDB Molecule:cellulose 1,4-beta-cellobiosidase;
PDBTitle: crystal structure of cellobiohydrolase from melanocarpus2 albomyces complexed with cellobiose
Phyre2
| 6 |
|
PDB 1ojj chain A
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 16.5%
Identity: 40%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Glycosyl hydrolase family 7 catalytic core
Phyre2
| 7 |
|
PDB 3ovw chain A
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 14.6%
Identity: 50%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Glycosyl hydrolase family 7 catalytic core
Phyre2
| 8 |
|
PDB 1eg1 chain A
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 14.0%
Identity: 50%
Fold: Concanavalin A-like lectins/glucanases
Superfamily: Concanavalin A-like lectins/glucanases
Family: Glycosyl hydrolase family 7 catalytic core
Phyre2
| 9 |
|
PDB 2j88 chain H domain 1
Region: 16 - 29
Aligned: 12
Modelled: 14
Confidence: 13.8%
Identity: 67%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: C1 set domains (antibody constant domain-like)
Phyre2
| 10 |
|
PDB 1v54 chain J
Region: 1 - 20
Aligned: 20
Modelled: 20
Confidence: 13.4%
Identity: 20%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIa
Family: Mitochondrial cytochrome c oxidase subunit VIIa
Phyre2
| 11 |
|
PDB 2a7o chain A
Region: 23 - 33
Aligned: 11
Modelled: 11
Confidence: 11.8%
Identity: 36%
PDB header:transcription
Chain: A: PDB Molecule:huntingtin interacting protein b;
PDBTitle: solution structure of the hset2/hypb sri domain
Phyre2
| 12 |
|
PDB 2yf3 chain F
Region: 22 - 31
Aligned: 10
Modelled: 10
Confidence: 11.7%
Identity: 30%
PDB header:hydrolase
Chain: F: PDB Molecule:mazg-like nucleoside triphosphate pyrophosphohydrolase;
PDBTitle: crystal structure of dr2231, the mazg-like protein from2 deinococcus radiodurans, complex with manganese
Phyre2
| 13 |
|
PDB 3cxb chain A
Region: 12 - 36
Aligned: 24
Modelled: 25
Confidence: 11.6%
Identity: 38%
PDB header:signaling protein
Chain: A: PDB Molecule:protein sifa;
PDBTitle: crystal structure of sifa and skip
Phyre2
| 14 |
|
PDB 2y69 chain W
Region: 1 - 20
Aligned: 20
Modelled: 20
Confidence: 11.1%
Identity: 20%
PDB header:electron transport
Chain: W: PDB Molecule:cytochrome c oxidase polypeptide 7a1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 15 |
|
PDB 1j3b chain A domain 1
Region: 15 - 21
Aligned: 7
Modelled: 7
Confidence: 9.3%
Identity: 57%
Fold: PEP carboxykinase-like
Superfamily: PEP carboxykinase-like
Family: PEP carboxykinase C-terminal domain
Phyre2
| 16 |
|
PDB 2fun chain B
Region: 20 - 31
Aligned: 12
Modelled: 12
Confidence: 8.6%
Identity: 33%
PDB header:apoptosis/hydrolase
Chain: B: PDB Molecule:caspase-8;
PDBTitle: alternative p35-caspase-8 complex
Phyre2
| 17 |
|
PDB 2h51 chain B
Region: 19 - 30
Aligned: 12
Modelled: 12
Confidence: 8.0%
Identity: 25%
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:caspase-1;
PDBTitle: crystal structure of human caspase-1 (glu390->asp and arg286->lys) in2 complex with 3-[2-(2-benzyloxycarbonylamino-3-methyl-butyrylamino)-3 propionylamino]-4-oxo-pentanoic acid (z-vad-fmk)
Phyre2
| 18 |
|
PDB 3o7v chain X
Region: 20 - 31
Aligned: 12
Modelled: 12
Confidence: 7.2%
Identity: 33%
PDB header:rna binding protein/rna
Chain: X: PDB Molecule:piwi-like protein 1;
PDBTitle: crystal structure of human hiwi1 (v361m) paz domain (residues 277-399)2 in complex with 14-mer rna (12-bp + 2-nt overhang) containing 2'-och33 at its 3'-end
Phyre2
| 19 |
|
PDB 3o7x chain C
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 7.1%
Identity: 33%
PDB header:rna binding protein
Chain: C: PDB Molecule:piwi-like protein 2;
PDBTitle: crystal structure of human hili paz domain
Phyre2
| 20 |
|
PDB 3ph0 chain C
Region: 1 - 16
Aligned: 16
Modelled: 16
Confidence: 6.3%
Identity: 25%
PDB header:chaperone
Chain: C: PDB Molecule:ascg;
PDBTitle: crystal structure of the heteromolecular chaperone, asce-ascg, from2 the type iii secretion system in aeromonas hydrophila
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|