| 1 |
|
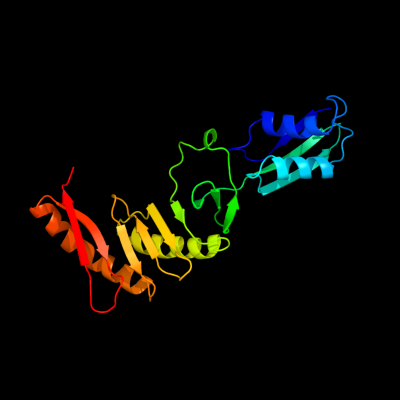
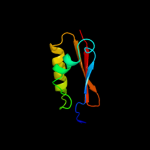
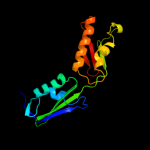
PDB 2vh1 chain A
Region: 58 - 260
Aligned: 203
Modelled: 203
Confidence: 100.0%
Identity: 100%
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsq;
PDBTitle: crystal structure of bacterial cell division protein ftsq2 from e.coli
Phyre2
| 2 |
|
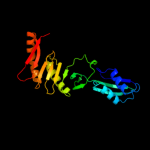
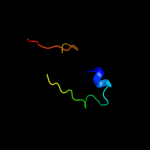
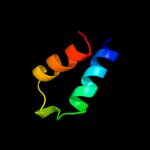
PDB 2vh2 chain A
Region: 56 - 260
Aligned: 200
Modelled: 205
Confidence: 100.0%
Identity: 72%
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsq;
PDBTitle: crystal structure of cell divison protein ftsq from2 yersinia enterecolitica
Phyre2
| 3 |
|
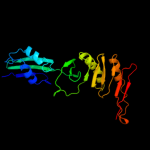
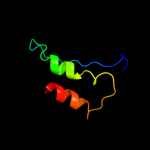
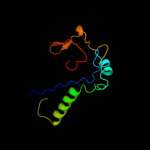
PDB 3j00 chain Z
Region: 22 - 116
Aligned: 94
Modelled: 94
Confidence: 99.8%
Identity: 89%
PDB header:ribosome/ribosomal protein
Chain: Z: PDB Molecule:cell division protein ftsq;
PDBTitle: structure of the ribosome-secye complex in the membrane environment
Phyre2
| 4 |
|
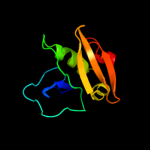
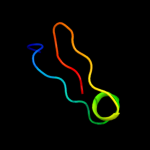
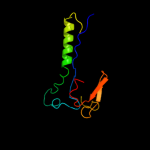
PDB 2alj chain A
Region: 124 - 221
Aligned: 97
Modelled: 98
Confidence: 98.6%
Identity: 12%
PDB header:cell cycle
Chain: A: PDB Molecule:cell-division initiation protein;
PDBTitle: structure of the cis confomer of the major extracytoplasmic2 domain of the bacterial cell division protein divib from3 geobacillus stearothermophilus
Phyre2
| 5 |
|
PDB 2qcz chain A
Region: 55 - 132
Aligned: 74
Modelled: 78
Confidence: 94.0%
Identity: 14%
PDB header:membrane protein, protein transport
Chain: A: PDB Molecule:outer membrane protein assembly factor yaet;
PDBTitle: structure of n-terminal domain of e. coli yaet
Phyre2
| 6 |
|
PDB 3efc chain A
Region: 55 - 132
Aligned: 74
Modelled: 78
Confidence: 92.6%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane protein assembly factor yaet;
PDBTitle: crystal structure of yaet periplasmic domain
Phyre2
| 7 |
|
PDB 2x8x chain X
Region: 53 - 128
Aligned: 73
Modelled: 76
Confidence: 79.3%
Identity: 12%
PDB header:chaperone
Chain: X: PDB Molecule:tlr1789 protein;
PDBTitle: structure of the n-terminal domain of omp85 from the2 thermophilic cyanobacterium thermosynechococcus elongatus
Phyre2
| 8 |
|
PDB 2v9h chain A
Region: 52 - 132
Aligned: 77
Modelled: 81
Confidence: 61.4%
Identity: 14%
PDB header:protein-binding
Chain: A: PDB Molecule:outer membrane protein assembly factor yaet;
PDBTitle: solution structure of an escherichia coli yaet tandem potra2 domain
Phyre2
| 9 |
|
PDB 3og5 chain A
Region: 49 - 126
Aligned: 75
Modelled: 78
Confidence: 52.2%
Identity: 16%
PDB header:protein binding
Chain: A: PDB Molecule:outer membrane protein assembly complex, yaet protein;
PDBTitle: crystal structure of bama potra45 tandem
Phyre2
| 10 |
|
PDB 3f41 chain B
Region: 227 - 259
Aligned: 33
Modelled: 33
Confidence: 39.2%
Identity: 27%
PDB header:hydrolase
Chain: B: PDB Molecule:phytase;
PDBTitle: structure of the tandemly repeated protein tyrosine2 phosphatase like phytase from mitsuokella multacida
Phyre2
| 11 |
|
PDB 2qdz chain A
Region: 53 - 101
Aligned: 47
Modelled: 49
Confidence: 20.4%
Identity: 4%
PDB header:protein transport
Chain: A: PDB Molecule:tpsb transporter fhac;
PDBTitle: structure of the membrane protein fhac: a member of the2 omp85/tpsb transporter family
Phyre2
| 12 |
|
PDB 2cw5 chain B
Region: 115 - 138
Aligned: 24
Modelled: 24
Confidence: 13.4%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:bacterial fluorinating enzyme homolog;
PDBTitle: crystal structure of a conserved hypothetical protein from2 thermus thermophilus hb8
Phyre2
| 13 |
|
PDB 3mc8 chain A
Region: 51 - 206
Aligned: 153
Modelled: 156
Confidence: 12.7%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:alr2269 protein;
PDBTitle: potra1-3 of the periplasmic domain of omp85 from anabaena
Phyre2
| 14 |
|
PDB 1q1v chain A
Region: 70 - 104
Aligned: 35
Modelled: 35
Confidence: 12.5%
Identity: 11%
Fold: Another 3-helical bundle
Superfamily: DEK C-terminal domain
Family: DEK C-terminal domain
Phyre2
| 15 |
|
PDB 3lno chain A
Region: 92 - 115
Aligned: 24
Modelled: 24
Confidence: 10.6%
Identity: 17%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
Phyre2
| 16 |
|
PDB 3jru chain B
Region: 125 - 220
Aligned: 88
Modelled: 96
Confidence: 8.5%
Identity: 11%
PDB header:hydrolase
Chain: B: PDB Molecule:probable cytosol aminopeptidase;
PDBTitle: crystal structure of leucyl aminopeptidase (pepa) from xoo0834,2 xanthomonas oryzae pv. oryzae kacc10331
Phyre2
| 17 |
|
PDB 1gyt chain A domain 2
Region: 125 - 220
Aligned: 88
Modelled: 96
Confidence: 7.6%
Identity: 15%
Fold: Phosphorylase/hydrolase-like
Superfamily: Zn-dependent exopeptidases
Family: Leucine aminopeptidase, C-terminal domain
Phyre2
| 18 |
|
PDB 3m92 chain B
Region: 122 - 142
Aligned: 21
Modelled: 21
Confidence: 6.6%
Identity: 10%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein ycin;
PDBTitle: the structure of ycin, an unchracterized protein from shigella2 flexneri.
Phyre2
| 19 |
|
PDB 1uwd chain A
Region: 92 - 115
Aligned: 23
Modelled: 24
Confidence: 5.4%
Identity: 13%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Fe-S cluster assembly (FSCA) domain-like
Family: PaaD-like
Phyre2