| 1 |
|
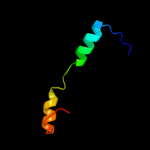
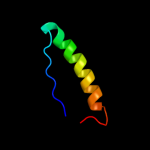
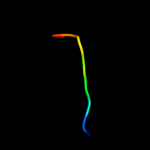
PDB 2knu chain A
Region: 70 - 81
Aligned: 12
Modelled: 12
Confidence: 47.3%
Identity: 50%
PDB header:membrane protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: solution structure of the transmembrane proximal region of2 the hepatis c virus e1 glycoprotein
Phyre2
| 2 |
|
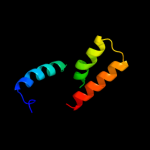
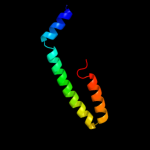
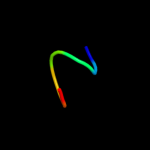
PDB 1lj2 chain B
Region: 6 - 53
Aligned: 41
Modelled: 48
Confidence: 32.4%
Identity: 27%
PDB header:viral protein/ translation
Chain: B: PDB Molecule:nonstructural rna-binding protein 34;
PDBTitle: recognition of eif4g by rotavirus nsp3 reveals a basis for2 mrna circularization
Phyre2
| 3 |
|
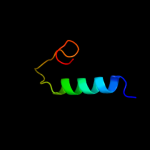
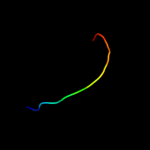
PDB 2jtv chain A
Region: 1 - 32
Aligned: 32
Modelled: 32
Confidence: 14.4%
Identity: 25%
PDB header:structural genomics
Chain: A: PDB Molecule:protein of unknown function;
PDBTitle: solution structure of protein rpa3401, northeast structural genomics2 consortium target rpt7, ontario center for structural proteomics3 target rp3384
Phyre2
| 4 |
|
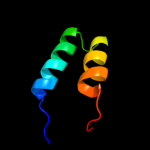
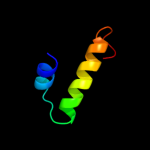
PDB 2ekw chain C
Region: 2 - 51
Aligned: 49
Modelled: 50
Confidence: 12.9%
Identity: 22%
PDB header:contractile protein
Chain: C: PDB Molecule:myosin catalytic light chain lc-1, mantle muscle;
PDBTitle: the crystal structure of squid myosin s1 in the presence of2 so4 2-
Phyre2
| 5 |
|
PDB 1wwi chain A domain 1
Region: 36 - 74
Aligned: 39
Modelled: 39
Confidence: 12.9%
Identity: 26%
Fold: Histone-fold
Superfamily: Histone-fold
Family: Bacterial histone-fold protein
Phyre2
| 6 |
|
PDB 1dip chain A
Region: 7 - 29
Aligned: 23
Modelled: 23
Confidence: 11.0%
Identity: 35%
PDB header:acetylation
Chain: A: PDB Molecule:delta-sleep-inducing peptide immunoreactive
PDBTitle: the solution structure of porcine delta-sleep-inducing2 peptide immunoreactive peptide, nmr, 10 structures
Phyre2
| 7 |
|
PDB 1r4v chain A
Region: 36 - 74
Aligned: 39
Modelled: 39
Confidence: 9.5%
Identity: 18%
Fold: Histone-fold
Superfamily: Histone-fold
Family: Bacterial histone-fold protein
Phyre2
| 8 |
|
PDB 1sl7 chain A
Region: 2 - 59
Aligned: 58
Modelled: 58
Confidence: 9.0%
Identity: 26%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Calmodulin-like
Phyre2
| 9 |
|
PDB 1fxk chain A
Region: 7 - 23
Aligned: 17
Modelled: 17
Confidence: 8.4%
Identity: 35%
Fold: Long alpha-hairpin
Superfamily: Prefoldin
Family: Prefoldin
Phyre2
| 10 |
|
PDB 1sfu chain A
Region: 3 - 34
Aligned: 30
Modelled: 32
Confidence: 8.0%
Identity: 23%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Z-DNA binding domain
Phyre2
| 11 |
|
PDB 2fm9 chain A domain 1
Region: 22 - 34
Aligned: 13
Modelled: 13
Confidence: 7.6%
Identity: 15%
Fold: SipA N-terminal domain-like
Superfamily: SipA N-terminal domain-like
Family: SipA N-terminal domain-like
Phyre2
| 12 |
|
PDB 1d1d chain A domain 2
Region: 3 - 42
Aligned: 39
Modelled: 40
Confidence: 6.6%
Identity: 26%
Fold: Retrovirus capsid protein, N-terminal core domain
Superfamily: Retrovirus capsid protein, N-terminal core domain
Family: Retrovirus capsid protein, N-terminal core domain
Phyre2
| 13 |
|
PDB 2gff chain B
Region: 33 - 66
Aligned: 34
Modelled: 34
Confidence: 5.9%
Identity: 12%
PDB header:sugar binding protein
Chain: B: PDB Molecule:lsrg protein;
PDBTitle: crystal structure of yersinia pestis lsrg
Phyre2
| 14 |
|
PDB 2ogd chain B
Region: 5 - 63
Aligned: 59
Modelled: 59
Confidence: 5.9%
Identity: 12%
PDB header:transferase
Chain: B: PDB Molecule:farnesyl pyrophosphate synthase;
PDBTitle: t. brucei farnesyl diphosphate synthase complexed with bisphosphonate2 bph-527
Phyre2
| 15 |
|
PDB 1z8y chain F
Region: 87 - 98
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 50%
PDB header:virus
Chain: F: PDB Molecule:spike glycoprotein e1;
PDBTitle: mapping the e2 glycoprotein of alphaviruses
Phyre2
| 16 |
|
PDB 1rj9 chain A domain 1
Region: 12 - 47
Aligned: 33
Modelled: 36
Confidence: 5.7%
Identity: 39%
Fold: Four-helical up-and-down bundle
Superfamily: Domain of the SRP/SRP receptor G-proteins
Family: Domain of the SRP/SRP receptor G-proteins
Phyre2
| 17 |
|
PDB 2v33 chain A domain 1
Region: 90 - 98
Aligned: 9
Modelled: 9
Confidence: 5.4%
Identity: 56%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: Class II viral fusion proteins C-terminal domain
Phyre2
| 18 |
|
PDB 2gyc chain 3 domain 1
Region: 51 - 57
Aligned: 7
Modelled: 7
Confidence: 5.4%
Identity: 57%
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Phyre2
| 19 |
|
PDB 1uiy chain A
Region: 9 - 55
Aligned: 45
Modelled: 47
Confidence: 5.3%
Identity: 29%
Fold: ClpP/crotonase
Superfamily: ClpP/crotonase
Family: Crotonase-like
Phyre2