| 1 |
|
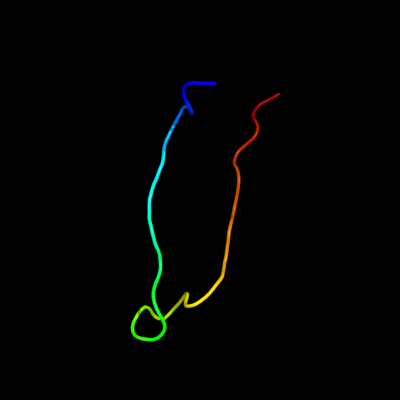
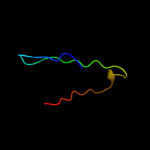
PDB 2wy3 chain B
Region: 29 - 53
Aligned: 21
Modelled: 25
Confidence: 29.3%
Identity: 29%
PDB header:immune system/viral protein
Chain: B: PDB Molecule:uncharacterized protein ul16;
PDBTitle: structure of the hcmv ul16-micb complex elucidates select2 binding of a viral immunoevasin to diverse nkg2d ligands
Phyre2
| 2 |
|
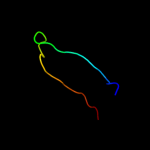
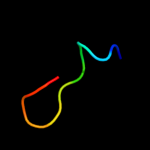
PDB 2o01 chain I
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 29.2%
Identity: 41%
PDB header:photosynthesis
Chain: I: PDB Molecule:photosystem i reaction center subunit viii;
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 3 |
|
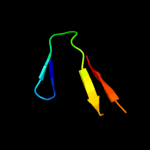
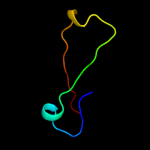
PDB 2rkz chain D
Region: 49 - 72
Aligned: 24
Modelled: 24
Confidence: 27.8%
Identity: 25%
PDB header:cell adhesion
Chain: D: PDB Molecule:fibronectin;
PDBTitle: crystal structure of the second and third fibronectin f12 modules in complex with a fragment of staphylococcus3 aureus fnbpa-1
Phyre2
| 4 |
|
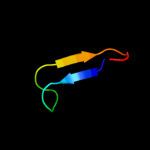
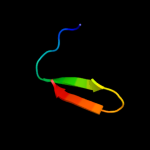
PDB 1zue chain A domain 1
Region: 43 - 51
Aligned: 9
Modelled: 9
Confidence: 25.6%
Identity: 33%
Fold: Defensin-like
Superfamily: Defensin-like
Family: Defensin
Phyre2
| 5 |
|
PDB 1nps chain A
Region: 47 - 68
Aligned: 22
Modelled: 22
Confidence: 21.1%
Identity: 14%
Fold: gamma-Crystallin-like
Superfamily: gamma-Crystallin-like
Family: Crystallins/Ca-binding development proteins
Phyre2
| 6 |
|
PDB 1wv3 chain A domain 2
Region: 53 - 74
Aligned: 21
Modelled: 22
Confidence: 19.4%
Identity: 43%
Fold: SMAD/FHA domain
Superfamily: SMAD/FHA domain
Family: EssC N-terminal domain-like
Phyre2
| 7 |
|
PDB 2fok chain A domain 4
Region: 50 - 59
Aligned: 10
Modelled: 10
Confidence: 17.7%
Identity: 30%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Restriction endonuclease FokI, C-terminal (catalytic) domain
Phyre2
| 8 |
|
PDB 1u5m chain A
Region: 49 - 66
Aligned: 18
Modelled: 18
Confidence: 16.5%
Identity: 39%
Fold: FnI-like domain
Superfamily: FnI-like domain
Family: VWC domain
Phyre2
| 9 |
|
PDB 1prs chain A domain 2
Region: 47 - 68
Aligned: 22
Modelled: 22
Confidence: 14.5%
Identity: 9%
Fold: gamma-Crystallin-like
Superfamily: gamma-Crystallin-like
Family: Crystallins/Ca-binding development proteins
Phyre2
| 10 |
|
PDB 3lw5 chain I
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 13.8%
Identity: 41%
PDB header:photosynthesis
Chain: I: PDB Molecule:photosystem i reaction center subunit viii;
PDBTitle: improved model of plant photosystem i
Phyre2
| 11 |
|
PDB 2wse chain I
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 13.8%
Identity: 41%
PDB header:photosynthesis
Chain: I: PDB Molecule:photosystem i reaction center subunit viii;
PDBTitle: improved model of plant photosystem i
Phyre2
| 12 |
|
PDB 1wv3 chain A
Region: 53 - 74
Aligned: 21
Modelled: 22
Confidence: 13.6%
Identity: 43%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:similar to dna segregation atpase and related
PDBTitle: crystal structure of n-terminal domain of hypothetical2 protein sav0287 from staphylococcus aureus
Phyre2
| 13 |
|
PDB 1hez chain D
Region: 56 - 90
Aligned: 35
Modelled: 35
Confidence: 10.1%
Identity: 29%
PDB header:antibody
Chain: D: PDB Molecule:heavy chain of ig;
PDBTitle: antibody-antigen complex
Phyre2
| 14 |
|
PDB 2vky chain B
Region: 23 - 36
Aligned: 14
Modelled: 14
Confidence: 9.2%
Identity: 29%
PDB header:viral protein
Chain: B: PDB Molecule:tail protein, piigcn4;
PDBTitle: headbinding domain of phage p22 tailspike c-terminally2 fused to isoleucine zipper piigcn4 (chimera i)
Phyre2
| 15 |
|
PDB 3i9h chain B
Region: 46 - 68
Aligned: 23
Modelled: 23
Confidence: 8.3%
Identity: 26%
PDB header:metal binding protein
Chain: B: PDB Molecule:beta and gamma crystallin;
PDBTitle: crystal structure of a betagamma-crystallin domain from2 clostridium beijerinckii
Phyre2
| 16 |
|
PDB 3dvg chain B domain 1
Region: 65 - 90
Aligned: 26
Modelled: 26
Confidence: 7.5%
Identity: 23%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: V set domains (antibody variable domain-like)
Phyre2
| 17 |
|
PDB 2orm chain A
Region: 30 - 41
Aligned: 12
Modelled: 12
Confidence: 7.3%
Identity: 33%
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase hp0924;
PDBTitle: crystal structure of the 4-oxalocrotonate tautomerase homologue dmpi2 from helicobacter pylori.
Phyre2
| 18 |
|
PDB 1adq chain H domain 1
Region: 57 - 90
Aligned: 34
Modelled: 34
Confidence: 6.5%
Identity: 24%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: V set domains (antibody variable domain-like)
Phyre2
| 19 |
|
PDB 1ed7 chain A
Region: 53 - 71
Aligned: 19
Modelled: 19
Confidence: 6.3%
Identity: 21%
Fold: WW domain-like
Superfamily: Carbohydrate binding domain
Family: Carbohydrate binding domain
Phyre2
| 20 |
|
PDB 2ko5 chain A
Region: 47 - 55
Aligned: 9
Modelled: 9
Confidence: 5.9%
Identity: 11%
PDB header:transcription
Chain: A: PDB Molecule:ring finger protein z;
PDBTitle: nmr solution structure of lfv-z
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|