| 1 |
|
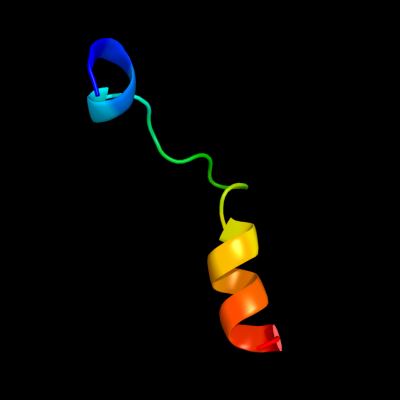
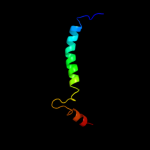
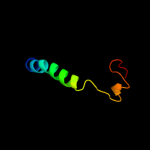
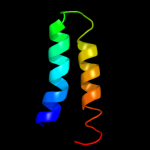
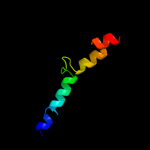
PDB 3qod chain B
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 42.0%
Identity: 35%
PDB header:dna binding protein
Chain: B: PDB Molecule:heterocyst differentiation protein;
PDBTitle: crystal structure of heterocyst differentiation protein, hetr from2 fischerella mv11
Phyre2
| 2 |
|
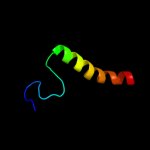
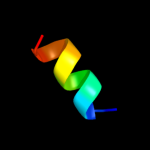
PDB 3qnq chain D
Region: 202 - 252
Aligned: 51
Modelled: 51
Confidence: 24.4%
Identity: 20%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 3 |
|
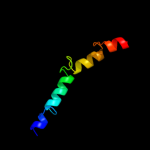
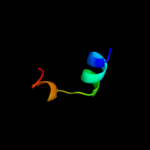
PDB 1v54 chain L
Region: 139 - 176
Aligned: 38
Modelled: 38
Confidence: 23.9%
Identity: 18%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Family: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Phyre2
| 4 |
|
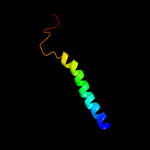
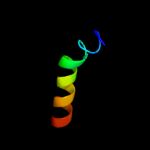
PDB 2klu chain A
Region: 207 - 258
Aligned: 52
Modelled: 52
Confidence: 19.8%
Identity: 19%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 5 |
|
PDB 2y69 chain Y
Region: 139 - 176
Aligned: 38
Modelled: 38
Confidence: 19.3%
Identity: 18%
PDB header:electron transport
Chain: Y: PDB Molecule:cytochrome c oxidase subunit 7c;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 6 |
|
PDB 2oar chain A domain 1
Region: 203 - 254
Aligned: 44
Modelled: 52
Confidence: 13.1%
Identity: 23%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 7 |
|
PDB 2knc chain A
Region: 212 - 251
Aligned: 40
Modelled: 40
Confidence: 10.8%
Identity: 13%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 8 |
|
PDB 2k21 chain A
Region: 216 - 258
Aligned: 43
Modelled: 43
Confidence: 10.2%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2
| 9 |
|
PDB 3h3p chain T
Region: 9 - 19
Aligned: 11
Modelled: 11
Confidence: 10.0%
Identity: 45%
PDB header:immune system
Chain: T: PDB Molecule:4e10_s0_1tjlc_004_n;
PDBTitle: crystal structure of hiv epitope-scaffold 4e10 fv complex
Phyre2
| 10 |
|
PDB 2aff chain B
Region: 241 - 258
Aligned: 18
Modelled: 18
Confidence: 8.9%
Identity: 17%
PDB header:cell cycle
Chain: B: PDB Molecule:mki67 fha domain interacting nucleolar phosphoprotein;
PDBTitle: the solution structure of the ki67fha/hnifk(226-269)3p complex
Phyre2
| 11 |
|
PDB 3j01 chain A
Region: 15 - 35
Aligned: 21
Modelled: 21
Confidence: 8.9%
Identity: 43%
PDB header:ribosome/ribosomal protein
Chain: A: PDB Molecule:preprotein translocase secy subunit;
PDBTitle: structure of the ribosome-secye complex in the membrane environment
Phyre2
| 12 |
|
PDB 2iub chain A domain 2
Region: 199 - 241
Aligned: 43
Modelled: 43
Confidence: 7.8%
Identity: 7%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 13 |
|
PDB 2rdd chain B
Region: 214 - 247
Aligned: 34
Modelled: 34
Confidence: 7.7%
Identity: 15%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 14 |
|
PDB 3iz6 chain C
Region: 236 - 257
Aligned: 22
Modelled: 22
Confidence: 7.5%
Identity: 18%
PDB header:ribosome
Chain: C: PDB Molecule:40s ribosomal protein s9 (s4p);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 15 |
|
PDB 2voy chain G
Region: 3 - 31
Aligned: 29
Modelled: 29
Confidence: 7.3%
Identity: 28%
PDB header:hydrolase
Chain: G: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 16 |
|
PDB 2jln chain A
Region: 151 - 257
Aligned: 103
Modelled: 107
Confidence: 6.6%
Identity: 16%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 17 |
|
PDB 2l0e chain A
Region: 201 - 219
Aligned: 19
Modelled: 19
Confidence: 5.2%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:sodium/hydrogen exchanger 1;
PDBTitle: structural and functional analysis of tm vi of the nhe1 isoform of the2 na+/h+ exchanger
Phyre2
| 18 |
|
PDB 2oar chain A
Region: 203 - 254
Aligned: 44
Modelled: 52
Confidence: 5.1%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2