| 1 |
|
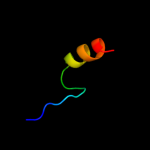
PDB 1u2z chain C
Region: 4 - 25
Aligned: 22
Modelled: 22
Confidence: 20.4%
Identity: 32%
PDB header:transferase
Chain: C: PDB Molecule:histone-lysine n-methyltransferase, h3 lysine-79
PDBTitle: crystal structure of histone k79 methyltransferase dot1p2 from yeast
Phyre2
| 2 |
|
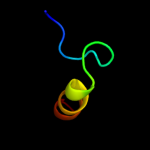
PDB 2iub chain A domain 2
Region: 6 - 25
Aligned: 20
Modelled: 20
Confidence: 9.4%
Identity: 30%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 3 |
|
PDB 2jm6 chain A
Region: 29 - 38
Aligned: 10
Modelled: 10
Confidence: 9.0%
Identity: 60%
PDB header:apoptosis
Chain: A: PDB Molecule:noxa;
PDBTitle: solution structure of mcl-1 complexed with noxab
Phyre2
| 4 |
|
PDB 2kbv chain A
Region: 17 - 24
Aligned: 8
Modelled: 8
Confidence: 8.9%
Identity: 75%
PDB header:membrane protein
Chain: A: PDB Molecule:sodium/hydrogen exchanger 1;
PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
Phyre2
| 5 |
|
PDB 1u2z chain A
Region: 4 - 25
Aligned: 22
Modelled: 22
Confidence: 8.5%
Identity: 32%
Fold: S-adenosyl-L-methionine-dependent methyltransferases
Superfamily: S-adenosyl-L-methionine-dependent methyltransferases
Family: Catalytic, N-terminal domain of histone methyltransferase Dot1l
Phyre2
| 6 |
|
PDB 3n4p chain A
Region: 45 - 51
Aligned: 7
Modelled: 7
Confidence: 6.2%
Identity: 71%
PDB header:dna binding protein
Chain: A: PDB Molecule:terminase subunit ul89 protein;
PDBTitle: human cytomegalovirus terminase nuclease domain
Phyre2
| 7 |
|
PDB 3n4q chain A
Region: 45 - 51
Aligned: 7
Modelled: 7
Confidence: 6.0%
Identity: 71%
PDB header:dna binding protein
Chain: A: PDB Molecule:terminase subunit ul89 protein;
PDBTitle: human cytomegalovirus terminase nuclease domain, mn soaked
Phyre2
| 8 |
|
PDB 2k5e chain A
Region: 2 - 22
Aligned: 21
Modelled: 21
Confidence: 5.5%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
Phyre2
| 9 |
|
PDB 2k53 chain A
Region: 2 - 22
Aligned: 21
Modelled: 21
Confidence: 5.4%
Identity: 33%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:a3dk08 protein;
PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
Phyre2
| 10 |
|
PDB 2l1n chain A
Region: 12 - 23
Aligned: 12
Modelled: 12
Confidence: 5.3%
Identity: 42%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the protein yp_399305.1
Phyre2