| 1 |
|
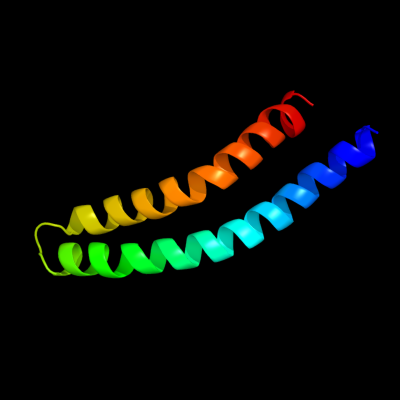
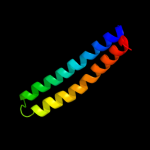
PDB 1c99 chain A
Region: 1 - 79
Aligned: 79
Modelled: 79
Confidence: 100.0%
Identity: 100%
Fold: Transmembrane helix hairpin
Superfamily: F1F0 ATP synthase subunit C
Family: F1F0 ATP synthase subunit C
Phyre2
| 2 |
|
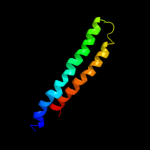
PDB 1wu0 chain A
Region: 6 - 75
Aligned: 70
Modelled: 70
Confidence: 99.9%
Identity: 44%
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase c chain;
PDBTitle: solution structure of subunit c of f1fo-atp synthase from2 the thermophilic bacillus ps3
Phyre2
| 3 |
|
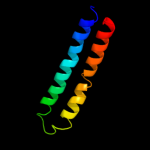
PDB 2x2v chain G
Region: 10 - 76
Aligned: 67
Modelled: 67
Confidence: 99.9%
Identity: 39%
PDB header:membrane protein
Chain: G: PDB Molecule:atp synthase subunit c;
PDBTitle: structural basis of a novel proton-coordination type in an2 f1fo-atp synthase rotor ring
Phyre2
| 4 |
|
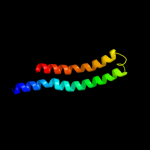
PDB 2w5j chain M
Region: 10 - 77
Aligned: 68
Modelled: 68
Confidence: 99.8%
Identity: 31%
PDB header:hydrolase
Chain: M: PDB Molecule:atp synthase c chain, chloroplastic;
PDBTitle: structure of the c14-rotor ring of the proton translocating2 chloroplast atp synthase
Phyre2
| 5 |
|
PDB 1yce chain D
Region: 10 - 77
Aligned: 67
Modelled: 68
Confidence: 99.8%
Identity: 24%
PDB header:membrane protein
Chain: D: PDB Molecule:subunit c;
PDBTitle: structure of the rotor ring of f-type na+-atpase from ilyobacter2 tartaricus
Phyre2
| 6 |
|
PDB 2wpd chain P
Region: 1 - 78
Aligned: 76
Modelled: 78
Confidence: 99.6%
Identity: 21%
PDB header:hydrolase
Chain: P: PDB Molecule:atp synthase subunit 9, mitochondrial;
PDBTitle: the mg.adp inhibited state of the yeast f1c10 atp synthase
Phyre2
| 7 |
|
PDB 2xnd chain K
Region: 10 - 76
Aligned: 66
Modelled: 67
Confidence: 99.5%
Identity: 23%
PDB header:hydrolase
Chain: K: PDB Molecule:atp synthase lipid-binding protein, mitochondrial;
PDBTitle: crystal structure of bovine f1-c8 sub-complex of atp2 synthase
Phyre2
| 8 |
|
PDB 2bl2 chain F
Region: 2 - 75
Aligned: 70
Modelled: 74
Confidence: 98.1%
Identity: 13%
PDB header:hydrolase
Chain: F: PDB Molecule:v-type sodium atp synthase subunit k;
PDBTitle: the membrane rotor of the v-type atpase from enterococcus2 hirae
Phyre2
| 9 |
|
PDB 2qqp chain D
Region: 24 - 42
Aligned: 19
Modelled: 19
Confidence: 12.5%
Identity: 47%
PDB header:virus
Chain: D: PDB Molecule:small capsid protein;
PDBTitle: crystal structure of authentic providence virus
Phyre2
| 10 |
|
PDB 1cf2 chain O domain 1
Region: 26 - 45
Aligned: 20
Modelled: 20
Confidence: 11.1%
Identity: 30%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 11 |
|
PDB 2voy chain D
Region: 10 - 25
Aligned: 16
Modelled: 16
Confidence: 7.9%
Identity: 50%
PDB header:hydrolase
Chain: D: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 12 |
|
PDB 1b7g chain O domain 1
Region: 26 - 45
Aligned: 20
Modelled: 20
Confidence: 7.7%
Identity: 30%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 13 |
|
PDB 1cf2 chain Q
Region: 26 - 45
Aligned: 20
Modelled: 20
Confidence: 7.4%
Identity: 30%
PDB header:oxidoreductase
Chain: Q: PDB Molecule:protein (glyceraldehyde-3-phosphate
PDBTitle: three-dimensional structure of d-glyceraldehyde-3-phosphate2 dehydrogenase from the hyperthermophilic archaeon3 methanothermus fervidus
Phyre2
| 14 |
|
PDB 1r0k chain A domain 2
Region: 26 - 44
Aligned: 19
Modelled: 19
Confidence: 6.2%
Identity: 32%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 15 |
|
PDB 2czc chain D
Region: 27 - 45
Aligned: 19
Modelled: 19
Confidence: 6.0%
Identity: 26%
PDB header:oxidoreductase
Chain: D: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 pyrococcus horikoshii ot3
Phyre2
| 16 |
|
PDB 2bby chain A
Region: 34 - 44
Aligned: 11
Modelled: 11
Confidence: 5.8%
Identity: 36%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: DNA-binding domain from rap30
Phyre2
| 17 |
|
PDB 1b7g chain O
Region: 27 - 45
Aligned: 19
Modelled: 19
Confidence: 5.4%
Identity: 32%
PDB header:oxidoreductase
Chain: O: PDB Molecule:protein (glyceraldehyde 3-phosphate dehydrogenase);
PDBTitle: glyceraldehyde 3-phosphate dehydrogenase
Phyre2
| 18 |
|
PDB 1vjq chain B
Region: 35 - 45
Aligned: 11
Modelled: 11
Confidence: 5.2%
Identity: 55%
PDB header:structural genomics, de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
Phyre2