| 1 |
|
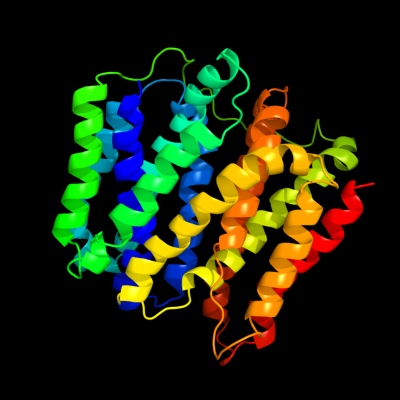
PDB 2gfp chain A
Region: 5 - 370
Aligned: 366
Modelled: 366
Confidence: 100.0%
Identity: 22%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 2 |
|
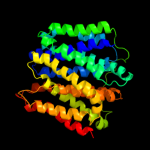
PDB 1pw4 chain A
Region: 3 - 390
Aligned: 387
Modelled: 388
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
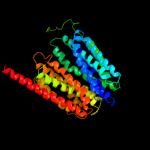
PDB 3o7p chain A
Region: 3 - 380
Aligned: 377
Modelled: 378
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
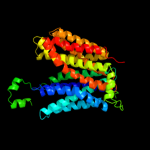
PDB 1pv7 chain A
Region: 2 - 389
Aligned: 388
Modelled: 388
Confidence: 100.0%
Identity: 9%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 7 - 375
Aligned: 369
Modelled: 369
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 2g9p chain A
Region: 57 - 70
Aligned: 14
Modelled: 14
Confidence: 20.6%
Identity: 36%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 7 |
|
PDB 2rdd chain B
Region: 362 - 390
Aligned: 29
Modelled: 29
Confidence: 7.4%
Identity: 21%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 8 |
|
PDB 3pro chain C domain 1
Region: 25 - 66
Aligned: 42
Modelled: 42
Confidence: 5.7%
Identity: 12%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2