| 1 |
|
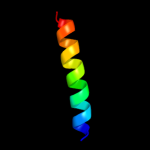
PDB 2lev chain A
Region: 47 - 66
Aligned: 20
Modelled: 20
Confidence: 13.4%
Identity: 25%
PDB header:transcription regulator/dna
Chain: A: PDB Molecule:ler;
PDBTitle: structure of the dna complex of the c-terminal domain of ler
Phyre2
| 2 |
|
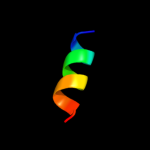
PDB 3fd9 chain C
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 10.3%
Identity: 19%
PDB header:unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the transcriptional anti-activator exsd2 from pseudomonas aeruginosa
Phyre2
| 3 |
|
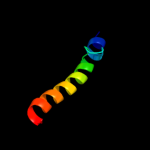
PDB 1t3t chain A domain 7
Region: 41 - 54
Aligned: 14
Modelled: 14
Confidence: 8.6%
Identity: 21%
Fold: PurM C-terminal domain-like
Superfamily: PurM C-terminal domain-like
Family: PurM C-terminal domain-like
Phyre2
| 4 |
|
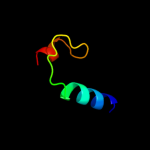
PDB 1vyt chain E
Region: 56 - 63
Aligned: 8
Modelled: 7
Confidence: 7.0%
Identity: 50%
PDB header:transport protein
Chain: E: PDB Molecule:voltage-dependent l-type calcium channel
PDBTitle: beta3 subunit complexed with aid
Phyre2
| 5 |
|
PDB 1rlj chain A
Region: 34 - 54
Aligned: 21
Modelled: 21
Confidence: 6.4%
Identity: 38%
Fold: Flavodoxin-like
Superfamily: Flavoproteins
Family: Flavoprotein NrdI
Phyre2
| 6 |
|
PDB 3thg chain A
Region: 28 - 49
Aligned: 22
Modelled: 22
Confidence: 6.4%
Identity: 32%
PDB header:protein binding
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase activase 1,
PDBTitle: crystal structure of the creosote rubisco activase c-domain
Phyre2
| 7 |
|
PDB 1x9b chain A
Region: 34 - 45
Aligned: 12
Modelled: 12
Confidence: 6.0%
Identity: 42%
Fold: Protozoan pheromone-like
Superfamily: Hypothetical membrane protein Ta0354, soluble domain
Family: Hypothetical membrane protein Ta0354, soluble domain
Phyre2
| 8 |
|
PDB 2knc chain A
Region: 8 - 40
Aligned: 29
Modelled: 33
Confidence: 5.7%
Identity: 24%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 9 |
|
PDB 2d0w chain A
Region: 35 - 64
Aligned: 30
Modelled: 30
Confidence: 5.6%
Identity: 7%
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome cl;
PDBTitle: crystal structure of cytochrome cl from hyphomicrobium2 denitrificans
Phyre2