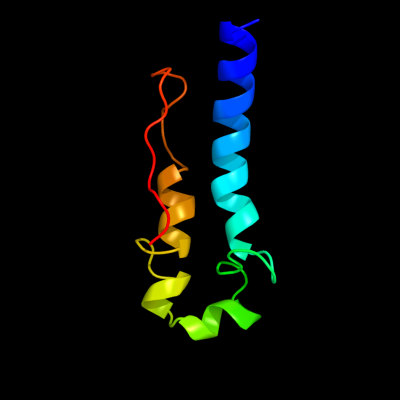
1 c2kz6A_
97.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of protein cv0426 from chromobacterium violaceum,2 northeast structural genomics consortium (nesg) target cvt2
2 c2jxwA_
69.3
34
PDB header: formin binding proteinChain: A: PDB Molecule: ww domain-binding protein 4;PDBTitle: solution structure of the tandem ww domains of fbp21
3 c2kt9A_
61.6
28
PDB header: ribosomal proteinChain: A: PDB Molecule: probable 30s ribosomal protein psrp-3;PDBTitle: solution nmr structure of probable 30s ribosomal protein2 psrp-3 (ycf65-like protein) from synechocystis sp. (strain3 pcc 6803), northeast structural genomics consortium target4 target sgr46
4 c1tk7A_
27.5
20
PDB header: signaling proteinChain: A: PDB Molecule: cg4244-pb;PDBTitle: nmr structure of ww domains (ww3-4) from suppressor of2 deltex
5 c2c7hA_
26.2
24
PDB header: ubiquitin-like proteinChain: A: PDB Molecule: retinoblastoma-binding protein 6, isoform 3;PDBTitle: solution nmr structure of the dwnn domain from human rbbp6
6 c1w18A_
23.9
20
PDB header: transferaseChain: A: PDB Molecule: levansucrase;PDBTitle: crystal structure of levansucrase from gluconacetobacter2 diazotrophicus
7 d1o6wa1
23.1
35
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain8 c2ov2O_
22.6
10
PDB header: protein binding/transferaseChain: O: PDB Molecule: serine/threonine-protein kinase pak 4;PDBTitle: the crystal structure of the human rac3 in complex with the crib2 domain of human p21-activated kinase 4 (pak4)
9 c1rfoC_
20.9
25
PDB header: viral proteinChain: C: PDB Molecule: whisker antigen control protein;PDBTitle: trimeric foldon of the t4 phagehead fibritin
10 c2odbB_
20.5
10
PDB header: protein bindingChain: B: PDB Molecule: serine/threonine-protein kinase pak 6;PDBTitle: the crystal structure of human cdc42 in complex with the crib domain2 of human p21-activated kinase 6 (pak6)
11 d1x4ta1
19.3
27
Fold: Long alpha-hairpinSuperfamily: ISY1 domain-likeFamily: ISY1 N-terminal domain-like12 d2ghsa1
17.2
16
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like13 c2ghsA_
17.2
16
PDB header: calcium-binding proteinChain: A: PDB Molecule: agr_c_1268p;PDBTitle: crystal structure of a calcium-binding protein, regucalcin2 (agr_c_1268) from agrobacterium tumefaciens str. c58 at 1.55 a3 resolution
14 c2xv5A_
16.1
13
PDB header: structural proteinChain: A: PDB Molecule: lamin-a/c;PDBTitle: human lamin a coil 2b fragment
15 c1e0aB_
14.7
30
PDB header: signalling proteinChain: B: PDB Molecule: serine/threonine-protein kinase pak-alpha;PDBTitle: cdc42 complexed with the gtpase binding domain of p212 activated kinase
16 c1gk6B_
14.4
14
PDB header: vimentinChain: B: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment linked to gcn4 leucine2 zipper (z2b)
17 c1zr7A_
14.2
39
PDB header: signaling proteinChain: A: PDB Molecule: huntingtin-interacting protein hypa/fbp11;PDBTitle: solution structure of the first ww domain of fbp11
18 c2dk7A_
14.1
21
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation regulator 1;PDBTitle: solution structure of ww domain in transcription elongation2 regulator 1
19 c2h47F_
13.6
19
PDB header: oxidoreductase/electron transportChain: F: PDB Molecule: aromatic amine dehydrogenase;PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
20 d1ll7a2
13.4
50
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain21 c1gq1B_
not modelled
13.0
14
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome cd1 nitrite reductase;PDBTitle: cytochrome cd1 nitrite reductase, y25s mutant, oxidised2 form
22 d1ln1a_
not modelled
12.3
19
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: STAR domain23 d2rm0w1
not modelled
12.3
33
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain24 c1f3mB_
not modelled
11.5
30
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase pak-alpha;PDBTitle: crystal structure of human serine/threonine kinase pak1
25 d1w9pa2
not modelled
11.4
42
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain26 c2diiA_
not modelled
11.0
7
PDB header: transcriptionChain: A: PDB Molecule: tfiih basal transcription factor complex p62PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
27 c1v1hB_
not modelled
10.6
25
PDB header: adenovirusChain: B: PDB Molecule: fibritin, fiber protein;PDBTitle: adenovirus fibre shaft sequence n-terminally fused to the2 bacteriophage t4 fibritin foldon trimerisation motif with3 a short linker
28 d2diia1
not modelled
10.6
7
Fold: BSD domain-likeSuperfamily: BSD domain-likeFamily: BSD domain29 c3o0rC_
not modelled
10.5
16
PDB header: immune system/oxidoreductaseChain: C: PDB Molecule: nitric oxide reductase subunit c;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
30 d1jofa_
not modelled
10.3
16
Fold: 7-bladed beta-propellerSuperfamily: 3-carboxy-cis,cis-mucoante lactonizing enzymeFamily: 3-carboxy-cis,cis-mucoante lactonizing enzyme31 d2dk1a1
not modelled
9.9
26
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain32 c1ox3A_
not modelled
9.9
25
PDB header: chaperoneChain: A: PDB Molecule: fibritin;PDBTitle: crystal structure of mini-fibritin
33 d1q67a_
not modelled
9.6
12
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Dcp134 c1x8yA_
not modelled
9.2
12
PDB header: structural proteinChain: A: PDB Molecule: lamin a/c;PDBTitle: human lamin coil 2b
35 d1pcna1
not modelled
9.0
47
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Colipase-likeFamily: Colipase-like36 d2e45a1
not modelled
8.7
27
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain37 c1avyA_
not modelled
8.7
25
PDB header: coiled coilChain: A: PDB Molecule: fibritin;PDBTitle: fibritin deletion mutant m (bacteriophage t4)
38 d2paja1
not modelled
8.7
12
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: SAH/MTA deaminase-like39 c2qkmG_
not modelled
8.6
35
PDB header: hydrolaseChain: G: PDB Molecule: spbc3b9.21 protein;PDBTitle: the crystal structure of fission yeast mrna decapping enzyme dcp1-dcp22 complex
40 c2dk1A_
not modelled
8.1
26
PDB header: gene regulationChain: A: PDB Molecule: ww domain-binding protein 4;PDBTitle: solution structure of ww domain in ww domain binding2 protein 4 (wbp-4)
41 c1gk4A_
not modelled
8.1
10
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment (cys2)
42 c1ymzA_
not modelled
7.8
27
PDB header: unknown functionChain: A: PDB Molecule: cc45;PDBTitle: cc45, an artificial ww domain designed using statistical2 coupling analysis
43 c2jz8A_
not modelled
7.8
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bh09830;PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
44 d2hqxa1
not modelled
7.7
20
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain45 c2hqxB_
not modelled
7.7
20
PDB header: transcriptionChain: B: PDB Molecule: p100 co-activator tudor domain;PDBTitle: crystal structure of human p100 tudor domain conserved2 region
46 d1pina1
not modelled
7.6
27
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain47 c2b9nZ_
not modelled
7.2
17
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein ctc;PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf2,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400.
48 c1nwdC_
not modelled
7.0
7
PDB header: binding protein/hydrolaseChain: C: PDB Molecule: glutamate decarboxylase;PDBTitle: solution structure of ca2+/calmodulin bound to the c-2 terminal domain of petunia glutamate decarboxylase
49 c1nwdB_
not modelled
7.0
7
PDB header: binding protein/hydrolaseChain: B: PDB Molecule: glutamate decarboxylase;PDBTitle: solution structure of ca2+/calmodulin bound to the c-2 terminal domain of petunia glutamate decarboxylase
50 c1q67B_
not modelled
6.8
12
PDB header: transcriptionChain: B: PDB Molecule: decapping protein involved in mrna degradation-PDBTitle: crystal structure of dcp1p
51 c3movB_
not modelled
6.7
13
PDB header: structural proteinChain: B: PDB Molecule: lamin-b1;PDBTitle: crystal structure of human lamin-b1 coil 2 segment
52 c2v43A_
not modelled
6.4
24
PDB header: regulatorChain: A: PDB Molecule: sigma-e factor regulatory protein rseb;PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
53 c2lawA_
not modelled
6.4
27
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: yorkie homolog;PDBTitle: structure of the second ww domain from human yap in complex with a2 human smad1 derived peptide
54 d1f8ab1
not modelled
6.4
27
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain55 d1o6wa2
not modelled
6.4
27
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain56 c1o6wA_
not modelled
6.3
35
PDB header: nuclear proteinChain: A: PDB Molecule: pre-mrna processing protein prp40;PDBTitle: solution structure of the prp40 ww domain pair of the yeast2 splicing factor prp40
57 c2ysiA_
not modelled
6.3
30
PDB header: protein bindingChain: A: PDB Molecule: transcription elongation regulator 1;PDBTitle: solution structure of the first ww domain from the mouse2 transcription elongation regulator 1, transcription factor3 ca150
58 c3gf8A_
not modelled
6.2
19
PDB header: carbohydrate binding proteinChain: A: PDB Molecule: putative polysaccharide binding proteins (duf1812);PDBTitle: crystal structure of putative polysaccharide binding proteins2 (duf1812) (np_809975.1) from bacteroides thetaiotaomicron vpi-5482 at3 2.20 a resolution
59 c1yiuA_
not modelled
6.2
23
PDB header: ligaseChain: A: PDB Molecule: itchy e3 ubiquitin protein ligase;PDBTitle: itch e3 ubiquitin ligase ww3 domain
60 d2madh_
not modelled
5.8
10
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain61 d1eysh1
not modelled
5.7
27
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: Photosynthetic reaction centre, H-chain, cytoplasmic domain62 c2i5nH_
not modelled
5.5
23
PDB header: photosynthesisChain: H: PDB Molecule: reaction center protein h chain;PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
63 c2kykA_
not modelled
5.4
27
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: the sandwich region between two lmp2a py motif regulates the2 interaction between aip4ww2domain and py motif
64 c1eysH_
not modelled
5.4
27
PDB header: electron transportChain: H: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
65 c2djyA_
not modelled
5.4
14
PDB header: ligase/signaling proteinChain: A: PDB Molecule: smad ubiquitination regulatory factor 2;PDBTitle: solution structure of smurf2 ww3 domain-smad7 py peptide2 complex
66 c2l5fA_
not modelled
5.2
33
PDB header: protein bindingChain: A: PDB Molecule: pre-mrna-processing factor 40 homolog a;PDBTitle: solution structure of the tandem ww domains from hypa/fbp11
67 d1d7qa_
not modelled
5.1
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like68 d2oa5a1
not modelled
5.0
21
Fold: BLRF2-likeSuperfamily: BLRF2-likeFamily: BLRF2-like69 d1zr7a1
not modelled
5.0
38
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain70 c3ok8A_
not modelled
5.0
0
PDB header: protein bindingChain: A: PDB Molecule: brain-specific angiogenesis inhibitor 1-associated proteinPDBTitle: i-bar of pinkbar
71 d1ywia1
not modelled
5.0
35
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain