| 1 |
|
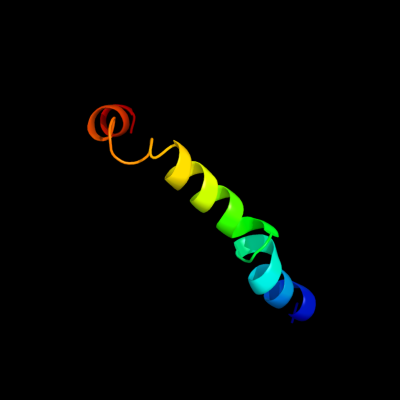
PDB 3rko chain F
Region: 41 - 83
Aligned: 42
Modelled: 43
Confidence: 21.7%
Identity: 14%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
PDB 1utc chain A domain 2
Region: 27 - 33
Aligned: 7
Modelled: 7
Confidence: 18.2%
Identity: 43%
Fold: 7-bladed beta-propeller
Superfamily: Clathrin heavy-chain terminal domain
Family: Clathrin heavy-chain terminal domain
Phyre2
| 3 |
|
PDB 1bpo chain A
Region: 27 - 33
Aligned: 7
Modelled: 7
Confidence: 17.9%
Identity: 43%
PDB header:membrane protein
Chain: A: PDB Molecule:protein (clathrin);
PDBTitle: clathrin heavy-chain terminal domain and linker
Phyre2
| 4 |
|
PDB 1c9l chain A
Region: 27 - 33
Aligned: 7
Modelled: 7
Confidence: 17.3%
Identity: 43%
PDB header:endocytosis/exocytosis
Chain: A: PDB Molecule:clathrin;
PDBTitle: peptide-in-groove interactions link target proteins to the2 b-propeller of clathrin
Phyre2
| 5 |
|
PDB 3qnq chain D
Region: 40 - 81
Aligned: 42
Modelled: 42
Confidence: 16.7%
Identity: 5%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 6 |
|
PDB 1xi4 chain D
Region: 27 - 33
Aligned: 7
Modelled: 7
Confidence: 13.7%
Identity: 43%
PDB header:endocytosis/exocytosis
Chain: D: PDB Molecule:clathrin heavy chain;
PDBTitle: clathrin d6 coat
Phyre2
| 7 |
|
PDB 2q04 chain C
Region: 23 - 34
Aligned: 12
Modelled: 12
Confidence: 10.8%
Identity: 25%
PDB header:transferase
Chain: C: PDB Molecule:acetoin utilization protein;
PDBTitle: crystal structure of acetoin utilization protein (zp_00540088.1) from2 exiguobacterium sibiricum 255-15 at 2.33 a resolution
Phyre2
| 8 |
|
PDB 2k59 chain B
Region: 12 - 29
Aligned: 18
Modelled: 18
Confidence: 6.6%
Identity: 28%
PDB header:transport protein
Chain: B: PDB Molecule:neuronal acetylcholine receptor subunit beta-2;
PDBTitle: nmr structures of the second transmembrane domain of the2 neuronal acetylcholine receptor beta 2 subunit
Phyre2
| 9 |
|
PDB 3c8e chain B
Region: 27 - 41
Aligned: 15
Modelled: 15
Confidence: 5.9%
Identity: 20%
PDB header:transferase
Chain: B: PDB Molecule:yghu, glutathione s-transferase homologue;
PDBTitle: crystal structure analysis of yghu from e. coli
Phyre2
| 10 |
|
PDB 1qhk chain A
Region: 26 - 37
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 25%
Fold: MbtH/L9 domain-like
Superfamily: L9 N-domain-like
Family: N-terminal domain of RNase HI
Phyre2
| 11 |
|
PDB 3bsu chain F
Region: 26 - 37
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 8%
PDB header:hydrolase/rna/dna
Chain: F: PDB Molecule:ribonuclease h1;
PDBTitle: hybrid-binding domain of human rnase h1 in complex with 12-2 mer rna/dna
Phyre2