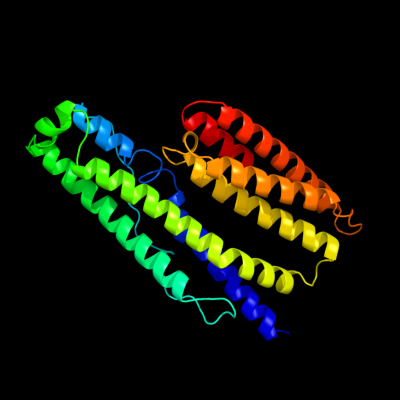
1 c3ieeA_
99.3
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative exported protein;PDBTitle: crystal structure of hypothetical protein bf3319 from bacteroides2 fragilis (yp_212931.1) from bacteroides fragilis nctc 9343 at 1.70 a3 resolution
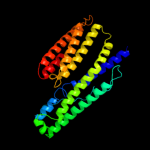
2 c3rh3A_
99.1
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized duf3829-like protein;PDBTitle: crystal structure of an uncharacterized duf3829-like protein (bt_1908)2 from bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution
3 d1vlta_
31.5
9
Fold: Four-helical up-and-down bundleSuperfamily: Aspartate receptor, ligand-binding domainFamily: Aspartate receptor, ligand-binding domain4 d1s94a_
21.1
16
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins5 c1s94A_
21.1
16
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: s-syntaxin;PDBTitle: crystal structure of the habc domain of neuronal syntaxin from the2 squid loligo pealei
6 c2kdtA_
15.8
33
PDB header: protein transportChain: A: PDB Molecule: neuroendocrine convertase 1;PDBTitle: pc1/3 dcsg sorting domain structure in dpc
7 c3ls1A_
15.2
13
PDB header: photosynthesisChain: A: PDB Molecule: sll1638 protein;PDBTitle: crystal structure of cyanobacterial psbq from synechocystis2 sp. pcc 6803 complexed with zn2+
8 c3c4mA_
13.7
15
PDB header: membrane proteinChain: A: PDB Molecule: fusion protein of maltose-binding periplasmic protein andPDBTitle: structure of human parathyroid hormone in complex with the2 extracellular domain of its g-protein-coupled receptor (pth1r)
9 c2kmfA_
12.8
17
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kda protein;PDBTitle: solution structure of psb27 from cyanobacterial photosystem2 ii
10 d1ykhb1
12.2
18
Fold: Mediator hinge subcomplex-likeSuperfamily: Mediator hinge subcomplex-likeFamily: CSE2-like11 c1pq1B_
12.2
27
PDB header: apoptosisChain: B: PDB Molecule: bcl2-like protein 11;PDBTitle: crystal structure of bcl-xl/bim
12 d1pgya_
12.0
60
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain13 c3fyqA_
11.7
13
PDB header: cell adhesionChain: A: PDB Molecule: cg6831-pa (talin);PDBTitle: structure of drosophila melanogaster talin ibs2 domain2 (residues 1981-2168)
14 d2liga_
11.3
9
Fold: Four-helical up-and-down bundleSuperfamily: Aspartate receptor, ligand-binding domainFamily: Aspartate receptor, ligand-binding domain15 d1ez3a_
11.3
10
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins16 d1sj8a2
10.0
17
Fold: I/LWEQ domainSuperfamily: I/LWEQ domainFamily: I/LWEQ domain17 c3d7vB_
9.1
22
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of mcl-1 in complex with an mcl-12 selective bh3 ligand
18 c2j1dG_
8.7
9
PDB header: protein bindingChain: G: PDB Molecule: disheveled-associated activator of morphogenesis 1;PDBTitle: crystallization of hdaam1 c-terminal fragment
19 d2jdia1
8.6
13
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase20 c2nl9B_
8.5
28
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: crystal structure of the mcl-1:bim bh3 complex
21 c3gvmA_
not modelled
8.0
12
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
22 d1wa8b1
not modelled
7.9
10
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like23 c3kj1B_
not modelled
7.9
38
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: mcl-1 in complex with bim bh3 mutant i2da
24 c2e76D_
not modelled
7.8
14
PDB header: photosynthesisChain: D: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
25 d1tuba2
not modelled
7.7
22
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain26 c2d4uA_
not modelled
7.4
12
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: crystal structure of the ligand binding domain of the bacterial serine2 chemoreceptor tsr
27 c2z6eC_
not modelled
6.9
8
PDB header: protein fibril regulatorChain: C: PDB Molecule: disheveled-associated activator of morphogenesisPDBTitle: crystal structure of human daam1 fh2
28 d1nm8a1
not modelled
6.9
20
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase29 d1hx1b_
not modelled
6.8
16
Fold: Spectrin repeat-likeSuperfamily: BAG domainFamily: BAG domain30 c3tekA_
not modelled
6.8
23
PDB header: dna binding proteinChain: A: PDB Molecule: thermodbp-single stranded dna binding protein;PDBTitle: thermodbp: a non-canonical single-stranded dna binding protein with a2 novel structure and mechanism
31 d1ndba1
not modelled
6.6
20
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase32 c2b8kD_
not modelled
6.6
17
PDB header: transferaseChain: D: PDB Molecule: dna-directed rna polymerase ii 32 kdaPDBTitle: 12-subunit rna polymerase ii
33 c2dkdA_
not modelled
6.5
22
PDB header: isomeraseChain: A: PDB Molecule: phosphoacetylglucosamine mutase;PDBTitle: crystal structure of n-acetylglucosamine-phosphate mutase,2 a member of the alpha-d-phosphohexomutase superfamily, in3 the product complex
34 d1v54e_
not modelled
6.5
20
Fold: alpha-alpha superhelixSuperfamily: Cytochrome c oxidase subunit EFamily: Cytochrome c oxidase subunit E35 c3kz0C_
not modelled
6.4
28
PDB header: apoptosisChain: C: PDB Molecule: mcl-1 specific peptide mb7;PDBTitle: mcl-1 complex with mcl-1-specific selected peptide
36 c3kz0D_
not modelled
6.4
28
PDB header: apoptosisChain: D: PDB Molecule: mcl-1 specific peptide mb7;PDBTitle: mcl-1 complex with mcl-1-specific selected peptide
37 c2vs0B_
not modelled
6.4
16
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
38 c2y69R_
not modelled
6.3
20
PDB header: electron transportChain: R: PDB Molecule: cytochrome c oxidase subunit 5a;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
39 d2f43a1
not modelled
6.1
12
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase40 c2k7wB_
not modelled
6.1
29
PDB header: apoptosisChain: B: PDB Molecule: bcl-2-like protein 11;PDBTitle: bax activation is initiated at a novel interaction site
41 c3s90B_
not modelled
6.1
9
PDB header: cell adhesionChain: B: PDB Molecule: vinculin;PDBTitle: human vinculin head domain vh1 (residues 1-252) in complex with murine2 talin (vbs33; residues 1512-1546)
42 c3s90A_
not modelled
6.1
9
PDB header: cell adhesionChain: A: PDB Molecule: vinculin;PDBTitle: human vinculin head domain vh1 (residues 1-252) in complex with murine2 talin (vbs33; residues 1512-1546)
43 c2qr4B_
not modelled
6.0
11
PDB header: hydrolaseChain: B: PDB Molecule: peptidase m3b, oligoendopeptidase f;PDBTitle: crystal structure of oligoendopeptidase-f from enterococcus faecium
44 d1skyb1
not modelled
6.0
12
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase45 c2jofA_
not modelled
5.9
50
PDB header: de novo proteinChain: A: PDB Molecule: trp-cage;PDBTitle: the trp-cage: optimizing the stability of a globular2 miniprotein
46 c1jrjA_
not modelled
5.8
7
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
47 c2npuA_
not modelled
5.8
17
PDB header: transferaseChain: A: PDB Molecule: fkbp12-rapamycin complex-associated protein;PDBTitle: the solution structure of the rapamycin-binding domain of2 mtor (frb)
48 d1tubb2
not modelled
5.8
22
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain49 d1u84a_
not modelled
5.7
29
Fold: YugE-likeSuperfamily: YugE-likeFamily: YugE-like50 d1fioa_
not modelled
5.6
10
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins51 d1p42a1
not modelled
5.6
16
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC52 d1fx0a1
not modelled
5.5
10
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase53 c2l81A_
not modelled
5.5
9
PDB header: cell adhesionChain: A: PDB Molecule: enhancer of filamentation 1;PDBTitle: solution nmr structure of the serine-rich domain of hef1 (enhancer of2 filamentation 1) from homo sapiens, northeast structural genomics3 consortium target hr5554a
54 c3mtuE_
not modelled
5.5
23
PDB header: contractile proteinChain: E: PDB Molecule: head morphogenesis protein, tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
55 c3tj5A_
not modelled
5.5
10
PDB header: protein binding/toxinChain: A: PDB Molecule: vinculin;PDBTitle: human vinculin head domain (vh1, residues 1-258) in complex with the2 vinculin binding site of the surface cell antigen 4 (sca4-vbs-n;3 residues 412-434) from rickettsia rickettsii
56 c1y9jA_
not modelled
5.3
23
PDB header: protein transportChain: A: PDB Molecule: sec1 family domain containing protein 1;PDBTitle: solution structure of the rat sly1 n-terminal domain