| 1 |
|
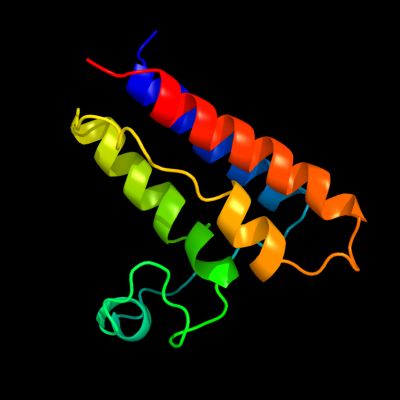
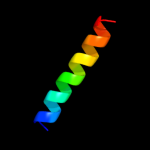
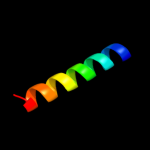
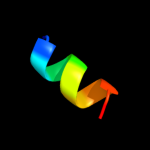
PDB 2hgk chain A domain 1
Region: 1 - 109
Aligned: 109
Modelled: 109
Confidence: 100.0%
Identity: 100%
Fold: Bromodomain-like
Superfamily: YqcC-like
Family: YqcC-like
Phyre2
| 2 |
|
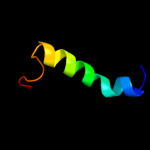
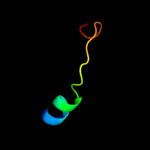
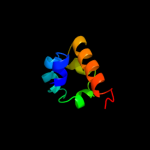
PDB 2gc6 chain A
Region: 45 - 64
Aligned: 20
Modelled: 20
Confidence: 31.5%
Identity: 30%
PDB header:ligase
Chain: A: PDB Molecule:folylpolyglutamate synthase;
PDBTitle: s73a mutant of l. casei fpgs
Phyre2
| 3 |
|
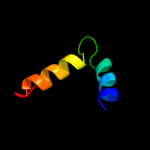
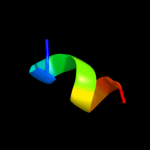
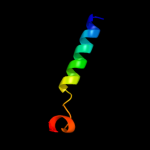
PDB 3d1d chain C
Region: 18 - 71
Aligned: 54
Modelled: 54
Confidence: 17.4%
Identity: 17%
PDB header:nuclear protein
Chain: C: PDB Molecule:rna-induced transcriptional silencing complex
PDBTitle: hexagonal crystal structure of tas3 c-terminal alpha motif
Phyre2
| 4 |
|
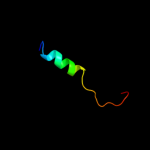
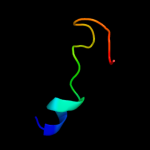
PDB 1ewf chain A domain 2
Region: 51 - 75
Aligned: 25
Modelled: 25
Confidence: 17.1%
Identity: 28%
Fold: Aha1/BPI domain-like
Superfamily: Bactericidal permeability-increasing protein, BPI
Family: Bactericidal permeability-increasing protein, BPI
Phyre2
| 5 |
|
PDB 1esw chain A
Region: 43 - 64
Aligned: 22
Modelled: 22
Confidence: 16.5%
Identity: 14%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Amylase, catalytic domain
Phyre2
| 6 |
|
PDB 1w1o chain A domain 1
Region: 2 - 29
Aligned: 28
Modelled: 28
Confidence: 15.0%
Identity: 18%
Fold: Ferredoxin-like
Superfamily: FAD-linked oxidases, C-terminal domain
Family: Cytokinin dehydrogenase 1
Phyre2
| 7 |
|
PDB 2yu0 chain A
Region: 71 - 103
Aligned: 32
Modelled: 33
Confidence: 11.1%
Identity: 19%
PDB header:signaling protein
Chain: A: PDB Molecule:interferon-activable protein 205;
PDBTitle: solution structures of the paad_dapin domain of mus2 musculus interferon-activatable protein 205
Phyre2
| 8 |
|
PDB 1bp1 chain A
Region: 51 - 75
Aligned: 25
Modelled: 25
Confidence: 9.7%
Identity: 28%
PDB header:bactericidal
Chain: A: PDB Molecule:bactericidal/permeability-increasing protein;
PDBTitle: crystal structure of bpi, the human bactericidal2 permeability-increasing protein
Phyre2
| 9 |
|
PDB 1x1n chain A domain 1
Region: 43 - 64
Aligned: 22
Modelled: 22
Confidence: 9.4%
Identity: 14%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Amylase, catalytic domain
Phyre2
| 10 |
|
PDB 2gc6 chain A domain 2
Region: 45 - 64
Aligned: 20
Modelled: 20
Confidence: 9.3%
Identity: 30%
Fold: Ribokinase-like
Superfamily: MurD-like peptide ligases, catalytic domain
Family: Folylpolyglutamate synthetase
Phyre2
| 11 |
|
PDB 1tz7 chain A domain 1
Region: 43 - 64
Aligned: 22
Modelled: 22
Confidence: 8.7%
Identity: 14%
Fold: TIM beta/alpha-barrel
Superfamily: (Trans)glycosidases
Family: Amylase, catalytic domain
Phyre2
| 12 |
|
PDB 1jb0 chain I
Region: 51 - 59
Aligned: 9
Modelled: 9
Confidence: 7.8%
Identity: 44%
Fold: Single transmembrane helix
Superfamily: Subunit VIII of photosystem I reaction centre, PsaI
Family: Subunit VIII of photosystem I reaction centre, PsaI
Phyre2
| 13 |
|
PDB 1pnb chain B
Region: 59 - 77
Aligned: 19
Modelled: 19
Confidence: 7.5%
Identity: 11%
PDB header:seed storage protein
Chain: B: PDB Molecule:napin bnib;
PDBTitle: structure of napin bnib, nmr, 10 structures
Phyre2
| 14 |
|
PDB 3if4 chain C
Region: 46 - 55
Aligned: 10
Modelled: 10
Confidence: 6.9%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:integron cassette protein hfx_cass5;
PDBTitle: structure from the mobile metagenome of north west arm2 sewage outfall: integron cassette protein hfx_cass5
Phyre2
| 15 |
|
PDB 1v38 chain A
Region: 48 - 109
Aligned: 61
Modelled: 62
Confidence: 6.1%
Identity: 13%
Fold: SAM domain-like
Superfamily: SAM/Pointed domain
Family: SAM (sterile alpha motif) domain
Phyre2
| 16 |
|
PDB 2r7g chain A domain 2
Region: 54 - 63
Aligned: 10
Modelled: 10
Confidence: 5.6%
Identity: 20%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Retinoblastoma tumor suppressor domains
Phyre2
| 17 |
|
PDB 3k1q chain A
Region: 40 - 64
Aligned: 24
Modelled: 25
Confidence: 5.6%
Identity: 13%
PDB header:virus
Chain: A: PDB Molecule:vp1, the protein that forms the mrna-capping
PDBTitle: backbone model of an aquareovirus virion by cryo-electron2 microscopy and bioinformatics
Phyre2
| 18 |
|
PDB 2fge chain A domain 3
Region: 9 - 36
Aligned: 28
Modelled: 28
Confidence: 5.3%
Identity: 14%
Fold: LuxS/MPP-like metallohydrolase
Superfamily: LuxS/MPP-like metallohydrolase
Family: MPP-like
Phyre2