| 1 |
|
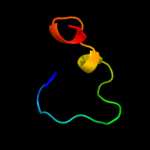
PDB 2aze chain A domain 1
Region: 330 - 339
Aligned: 10
Modelled: 10
Confidence: 28.4%
Identity: 30%
Fold: E2F-DP heterodimerization region
Superfamily: E2F-DP heterodimerization region
Family: DP dimerization segment
Phyre2
| 2 |
|
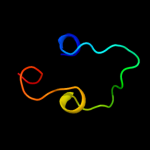
PDB 2l2t chain A
Region: 337 - 369
Aligned: 33
Modelled: 33
Confidence: 25.4%
Identity: 21%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 3 |
|
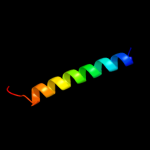
PDB 2knc chain B
Region: 272 - 299
Aligned: 28
Modelled: 28
Confidence: 23.8%
Identity: 29%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 4 |
|
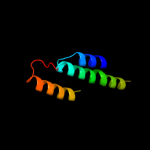
PDB 1iwg chain A domain 8
Region: 272 - 369
Aligned: 82
Modelled: 82
Confidence: 12.0%
Identity: 9%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 5 |
|
PDB 2jp3 chain A
Region: 3 - 37
Aligned: 35
Modelled: 35
Confidence: 11.7%
Identity: 14%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 6 |
|
PDB 2jo1 chain A
Region: 2 - 37
Aligned: 36
Modelled: 36
Confidence: 11.6%
Identity: 19%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 7 |
|
PDB 2gdw chain A domain 1
Region: 325 - 349
Aligned: 24
Modelled: 25
Confidence: 9.5%
Identity: 25%
Fold: Acyl carrier protein-like
Superfamily: ACP-like
Family: Peptidyl carrier domain
Phyre2
| 8 |
|
PDB 2png chain A domain 1
Region: 322 - 349
Aligned: 28
Modelled: 28
Confidence: 9.0%
Identity: 21%
Fold: Acyl carrier protein-like
Superfamily: ACP-like
Family: Acyl-carrier protein (ACP)
Phyre2
| 9 |
|
PDB 2rmz chain A
Region: 272 - 300
Aligned: 29
Modelled: 29
Confidence: 8.5%
Identity: 28%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin beta-3;
PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
Phyre2
| 10 |
|
PDB 1ywu chain A
Region: 1 - 17
Aligned: 17
Modelled: 17
Confidence: 6.5%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa4608;
PDBTitle: solution nmr structure of pseudomonas aeruginosa protein pa4608.2 northeast structural genomics target pat7
Phyre2
| 11 |
|
PDB 1ywu chain A domain 1
Region: 1 - 17
Aligned: 17
Modelled: 17
Confidence: 6.5%
Identity: 18%
Fold: Split barrel-like
Superfamily: PilZ domain-like
Family: PilZ domain
Phyre2
| 12 |
|
PDB 3rko chain K
Region: 305 - 369
Aligned: 64
Modelled: 65
Confidence: 5.2%
Identity: 19%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2