| 1 |
|
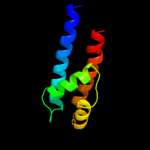
PDB 3pjz chain A
Region: 5 - 484
Aligned: 464
Modelled: 464
Confidence: 100.0%
Identity: 42%
PDB header:transport protein
Chain: A: PDB Molecule:potassium uptake protein trkh;
PDBTitle: crystal structure of the potassium transporter trkh from vibrio2 parahaemolyticus
Phyre2
| 2 |
|
PDB 3e8g chain B
Region: 272 - 346
Aligned: 69
Modelled: 75
Confidence: 94.2%
Identity: 16%
PDB header:membrane protein
Chain: B: PDB Molecule:potassium channel protein;
PDBTitle: crystal structure of the the open nak channel-na+/ca2+ complex
Phyre2
| 3 |
|
PDB 1f6g chain A
Region: 45 - 188
Aligned: 139
Modelled: 144
Confidence: 91.1%
Identity: 16%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 4 |
|
PDB 1p7b chain B
Region: 66 - 158
Aligned: 90
Modelled: 93
Confidence: 86.0%
Identity: 14%
PDB header:metal transport
Chain: B: PDB Molecule:integral membrane channel and cytosolic domains;
PDBTitle: crystal structure of an inward rectifier potassium channel
Phyre2
| 5 |
|
PDB 2qks chain A
Region: 66 - 162
Aligned: 94
Modelled: 97
Confidence: 85.4%
Identity: 16%
PDB header:metal transport
Chain: A: PDB Molecule:kir3.1-prokaryotic kir channel chimera;
PDBTitle: crystal structure of a kir3.1-prokaryotic kir channel chimera
Phyre2
| 6 |
|
PDB 1xl6 chain B
Region: 66 - 158
Aligned: 90
Modelled: 93
Confidence: 84.8%
Identity: 16%
PDB header:metal transport
Chain: B: PDB Molecule:inward rectifier potassium channel;
PDBTitle: intermediate gating structure 2 of the inwardly rectifying k+ channel2 kirbac3.1
Phyre2
| 7 |
|
PDB 3jyc chain A
Region: 66 - 158
Aligned: 90
Modelled: 93
Confidence: 82.4%
Identity: 14%
PDB header:metal transport
Chain: A: PDB Molecule:inward-rectifier k+ channel kir2.2;
PDBTitle: crystal structure of the eukaryotic strong inward-rectifier2 k+ channel kir2.2 at 3.1 angstrom resolution
Phyre2
| 8 |
|
PDB 3ifx chain B
Region: 397 - 467
Aligned: 65
Modelled: 71
Confidence: 73.0%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:voltage-gated potassium channel;
PDBTitle: crystal structure of the spin-labeled kcsa mutant v48r1
Phyre2
| 9 |
|
PDB 2kb1 chain A
Region: 422 - 467
Aligned: 40
Modelled: 46
Confidence: 65.1%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2
| 10 |
|
PDB 1p7b chain A domain 2
Region: 66 - 158
Aligned: 90
Modelled: 93
Confidence: 57.2%
Identity: 16%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 11 |
|
PDB 1xl4 chain A domain 2
Region: 66 - 153
Aligned: 84
Modelled: 88
Confidence: 54.9%
Identity: 13%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 12 |
|
PDB 1r3j chain C
Region: 397 - 467
Aligned: 65
Modelled: 71
Confidence: 51.5%
Identity: 11%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 13 |
|
PDB 1lnq chain C
Region: 402 - 467
Aligned: 60
Modelled: 66
Confidence: 50.0%
Identity: 12%
PDB header:metal transport
Chain: C: PDB Molecule:potassium channel related protein;
PDBTitle: crystal structure of mthk at 3.3 a
Phyre2
| 14 |
|
PDB 2a9h chain A domain 1
Region: 397 - 482
Aligned: 80
Modelled: 86
Confidence: 33.0%
Identity: 11%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 15 |
|
PDB 2r9r chain H
Region: 328 - 478
Aligned: 140
Modelled: 151
Confidence: 24.4%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: H: PDB Molecule:paddle chimera voltage gated potassium channel kv1.2-kv2.1;
PDBTitle: shaker family voltage dependent potassium channel (kv1.2-kv2.1 paddle2 chimera channel) in association with beta subunit
Phyre2
| 16 |
|
PDB 1lnq chain A domain 2
Region: 420 - 467
Aligned: 42
Modelled: 48
Confidence: 24.0%
Identity: 21%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 17 |
|
PDB 2h8p chain C domain 1
Region: 272 - 323
Aligned: 52
Modelled: 52
Confidence: 15.6%
Identity: 13%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 18 |
|
PDB 3beh chain A
Region: 392 - 476
Aligned: 78
Modelled: 85
Confidence: 12.4%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:mll3241 protein;
PDBTitle: structure of a bacterial cyclic nucleotide regulated ion channel
Phyre2