| 1 |
|
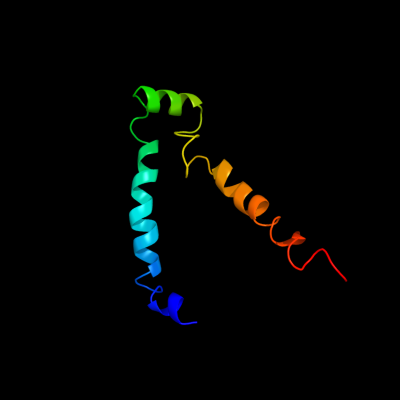
PDB 2aki chain Y
Region: 337 - 412
Aligned: 65
Modelled: 76
Confidence: 37.6%
Identity: 14%
PDB header:protein transport
Chain: Y: PDB Molecule:preprotein translocase secy subunit;
PDBTitle: normal mode-based flexible fitted coordinates of a2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
Phyre2
| 2 |
|
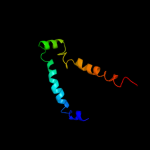
PDB 3dl8 chain H
Region: 337 - 412
Aligned: 65
Modelled: 76
Confidence: 28.5%
Identity: 9%
PDB header:protein transport
Chain: H: PDB Molecule:preprotein translocase subunit secy;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
Phyre2
| 3 |
|
PDB 2aze chain A domain 1
Region: 368 - 377
Aligned: 10
Modelled: 10
Confidence: 27.4%
Identity: 40%
Fold: E2F-DP heterodimerization region
Superfamily: E2F-DP heterodimerization region
Family: DP dimerization segment
Phyre2
| 4 |
|
PDB 2knc chain B
Region: 307 - 343
Aligned: 33
Modelled: 37
Confidence: 22.9%
Identity: 15%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 5 |
|
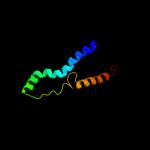
PDB 3j01 chain A
Region: 337 - 412
Aligned: 65
Modelled: 76
Confidence: 21.6%
Identity: 11%
PDB header:ribosome/ribosomal protein
Chain: A: PDB Molecule:preprotein translocase secy subunit;
PDBTitle: structure of the ribosome-secye complex in the membrane environment
Phyre2
| 6 |
|
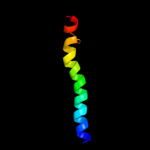
PDB 2klu chain A
Region: 373 - 410
Aligned: 38
Modelled: 38
Confidence: 17.3%
Identity: 18%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 7 |
|
PDB 2gdw chain A domain 1
Region: 363 - 387
Aligned: 24
Modelled: 25
Confidence: 14.0%
Identity: 29%
Fold: Acyl carrier protein-like
Superfamily: ACP-like
Family: Peptidyl carrier domain
Phyre2
| 8 |
|
PDB 3din chain F
Region: 337 - 412
Aligned: 65
Modelled: 76
Confidence: 12.4%
Identity: 14%
PDB header:membrane protein, protein transport
Chain: F: PDB Molecule:preprotein translocase subunit secy;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 9 |
|
PDB 2png chain A domain 1
Region: 360 - 387
Aligned: 28
Modelled: 28
Confidence: 10.9%
Identity: 29%
Fold: Acyl carrier protein-like
Superfamily: ACP-like
Family: Acyl-carrier protein (ACP)
Phyre2
| 10 |
|
PDB 1iwg chain A domain 8
Region: 349 - 411
Aligned: 63
Modelled: 63
Confidence: 7.5%
Identity: 16%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 11 |
|
PDB 2ywx chain A
Region: 367 - 389
Aligned: 22
Modelled: 23
Confidence: 5.6%
Identity: 18%
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
Phyre2
| 12 |
|
PDB 2cq8 chain A
Region: 363 - 390
Aligned: 28
Modelled: 28
Confidence: 5.3%
Identity: 7%
PDB header:oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: solution structure of rsgi ruh-033, a pp-binding domain of2 10-fthfdh from human cdna
Phyre2
| 13 |
|
PDB 1ar1 chain B
Region: 43 - 110
Aligned: 68
Modelled: 68
Confidence: 5.0%
Identity: 4%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2