| 1 |
|
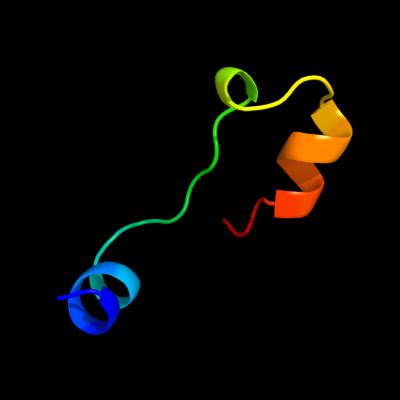
PDB 2v5i chain A
Region: 14 - 43
Aligned: 30
Modelled: 30
Confidence: 15.3%
Identity: 30%
PDB header:viral protein
Chain: A: PDB Molecule:salmonella typhimurium db7155 bacteriophage det7
PDBTitle: structure of the receptor-binding protein of bacteriophage2 det7: a podoviral tailspike in a myovirus
Phyre2
| 2 |
|
PDB 1uww chain A
Region: 24 - 30
Aligned: 7
Modelled: 7
Confidence: 12.1%
Identity: 71%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Family 28 carbohydrate binding module, CBM28
Phyre2
| 3 |
|
PDB 1tyw chain A
Region: 14 - 43
Aligned: 30
Modelled: 30
Confidence: 9.6%
Identity: 33%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pectin lyase-like
Family: P22 tailspike protein
Phyre2
| 4 |
|
PDB 3acg chain A
Region: 24 - 30
Aligned: 7
Modelled: 7
Confidence: 8.6%
Identity: 57%
PDB header:hydrolase
Chain: A: PDB Molecule:beta-1,4-endoglucanase;
PDBTitle: crystal structure of carbohydrate-binding module family 282 from clostridium josui cel5a in complex with cellobiose
Phyre2
| 5 |
|
PDB 2dla chain B
Region: 19 - 31
Aligned: 13
Modelled: 13
Confidence: 8.2%
Identity: 69%
PDB header:replication
Chain: B: PDB Molecule:397aa long hypothetical protein;
PDBTitle: primase large subunit amino terminal domain from pyrococcus horikoshii
Phyre2
| 6 |
|
PDB 2xc1 chain A
Region: 14 - 43
Aligned: 30
Modelled: 30
Confidence: 6.8%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:bifunctional tail protein;
PDBTitle: full-length tailspike protein mutant y108w of bacteriophage2 p22
Phyre2
| 7 |
|
PDB 3ech chain C
Region: 47 - 54
Aligned: 8
Modelled: 8
Confidence: 6.8%
Identity: 75%
PDB header:transcription, transcription regulation
Chain: C: PDB Molecule:25-mer fragment of protein armr;
PDBTitle: the marr-family repressor mexr in complex with its antirepressor armr
Phyre2
| 8 |
|
PDB 2wg3 chain C
Region: 10 - 41
Aligned: 32
Modelled: 32
Confidence: 6.7%
Identity: 34%
PDB header:signaling protein
Chain: C: PDB Molecule:hedgehog-interacting protein;
PDBTitle: crystal structure of the complex between human hedgehog-2 interacting protein hip and desert hedgehog without calcium
Phyre2
| 9 |
|
PDB 1ed7 chain A
Region: 10 - 34
Aligned: 25
Modelled: 25
Confidence: 5.8%
Identity: 28%
Fold: WW domain-like
Superfamily: Carbohydrate binding domain
Family: Carbohydrate binding domain
Phyre2