| 1 |
|
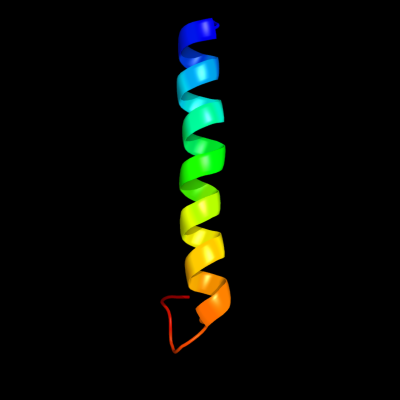
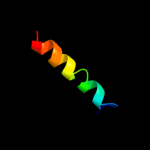
PDB 2l2t chain A
Region: 38 - 68
Aligned: 31
Modelled: 31
Confidence: 32.0%
Identity: 26%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 2 |
|
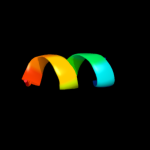
PDB 2qjx chain A
Region: 2 - 34
Aligned: 22
Modelled: 33
Confidence: 24.2%
Identity: 32%
PDB header:protein binding
Chain: A: PDB Molecule:protein bim1;
PDBTitle: structural basis of microtubule plus end tracking by2 xmap215, clip-170 and eb1
Phyre2
| 3 |
|
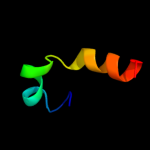
PDB 3kdp chain G
Region: 43 - 56
Aligned: 14
Modelled: 14
Confidence: 23.1%
Identity: 36%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 4 |
|
PDB 3kdp chain H
Region: 43 - 56
Aligned: 14
Modelled: 14
Confidence: 23.1%
Identity: 36%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 5 |
|
PDB 1wyo chain A
Region: 2 - 34
Aligned: 22
Modelled: 33
Confidence: 16.9%
Identity: 32%
PDB header:structural protein
Chain: A: PDB Molecule:microtubule-associated protein rp/eb family
PDBTitle: solution structure of the ch domain of human microtubule-2 associated protein rp/eb family member 3
Phyre2
| 6 |
|
PDB 2gbs chain A domain 1
Region: 31 - 45
Aligned: 15
Modelled: 15
Confidence: 11.3%
Identity: 33%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: Atu2648/PH1033-like
Phyre2
| 7 |
|
PDB 2qjz chain A domain 1
Region: 2 - 35
Aligned: 23
Modelled: 23
Confidence: 10.2%
Identity: 30%
Fold: CH domain-like
Superfamily: Calponin-homology domain, CH-domain
Family: Calponin-homology domain, CH-domain
Phyre2
| 8 |
|
PDB 3ixz chain B
Region: 32 - 53
Aligned: 22
Modelled: 22
Confidence: 10.1%
Identity: 18%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
Phyre2
| 9 |
|
PDB 3bq9 chain A
Region: 53 - 71
Aligned: 19
Modelled: 19
Confidence: 9.7%
Identity: 32%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
Phyre2
| 10 |
|
PDB 3o0r chain C
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 9.6%
Identity: 35%
PDB header:immune system/oxidoreductase
Chain: C: PDB Molecule:nitric oxide reductase subunit c;
PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
Phyre2
| 11 |
|
PDB 1m56 chain D
Region: 36 - 50
Aligned: 15
Modelled: 15
Confidence: 9.5%
Identity: 40%
Fold: Single transmembrane helix
Superfamily: Bacterial aa3 type cytochrome c oxidase subunit IV
Family: Bacterial aa3 type cytochrome c oxidase subunit IV
Phyre2
| 12 |
|
PDB 2kdn chain A
Region: 28 - 39
Aligned: 12
Modelled: 12
Confidence: 9.2%
Identity: 17%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein pfe0790c;
PDBTitle: solution structure of pfe0790c, a putative bola-like2 protein from the protozoan parasite plasmodium falciparum.
Phyre2
| 13 |
|
PDB 1v5k chain A
Region: 2 - 34
Aligned: 22
Modelled: 33
Confidence: 8.9%
Identity: 32%
Fold: CH domain-like
Superfamily: Calponin-homology domain, CH-domain
Family: Calponin-homology domain, CH-domain
Phyre2
| 14 |
|
PDB 3b8e chain B
Region: 32 - 53
Aligned: 22
Modelled: 22
Confidence: 8.8%
Identity: 23%
PDB header:hydrolase/transport protein
Chain: B: PDB Molecule:sodium/potassium-transporting atpase subunit
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 15 |
|
PDB 1eys chain H domain 2
Region: 56 - 65
Aligned: 10
Modelled: 10
Confidence: 8.7%
Identity: 40%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 16 |
|
PDB 3dl8 chain D
Region: 41 - 58
Aligned: 18
Modelled: 18
Confidence: 8.7%
Identity: 28%
PDB header:protein transport
Chain: D: PDB Molecule:sece;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
Phyre2
| 17 |
|
PDB 3o2e chain A
Region: 31 - 38
Aligned: 8
Modelled: 8
Confidence: 7.7%
Identity: 50%
PDB header:unknown function
Chain: A: PDB Molecule:bola-like protein;
PDBTitle: crystal structure of a bol-like protein from babesia bovis
Phyre2
| 18 |
|
PDB 2veq chain A
Region: 38 - 63
Aligned: 26
Modelled: 26
Confidence: 7.5%
Identity: 31%
PDB header:cell cycle
Chain: A: PDB Molecule:centromere dna-binding protein complex cbf3
PDBTitle: insights into kinetochore-dna interactions from the2 structure of cep3p
Phyre2
| 19 |
|
PDB 1ny8 chain A
Region: 31 - 39
Aligned: 9
Modelled: 9
Confidence: 7.3%
Identity: 44%
Fold: Alpha-lytic protease prodomain-like
Superfamily: BolA-like
Family: BolA-like
Phyre2
| 20 |
|
PDB 1v9j chain A
Region: 31 - 38
Aligned: 8
Modelled: 8
Confidence: 6.8%
Identity: 50%
Fold: Alpha-lytic protease prodomain-like
Superfamily: BolA-like
Family: BolA-like
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|