| 1 |
|
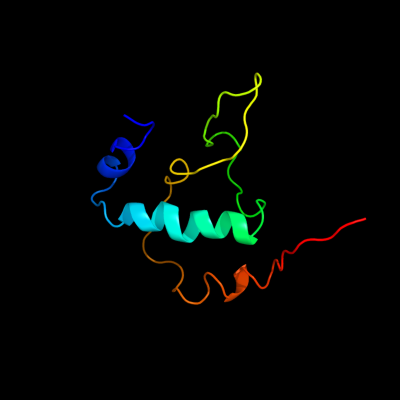
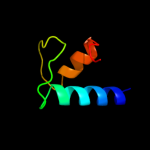
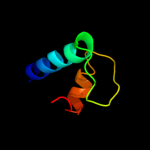
PDB 3iyd chain E
Region: 2 - 91
Aligned: 90
Modelled: 90
Confidence: 100.0%
Identity: 100%
PDB header:transcription/dna
Chain: E: PDB Molecule:dna-directed rna polymerase subunit omega;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
Phyre2
| 2 |
|
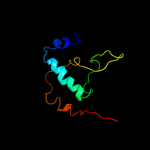
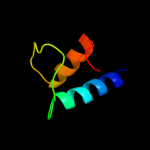
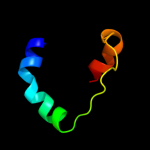
PDB 1ynj chain K domain 1
Region: 1 - 60
Aligned: 60
Modelled: 60
Confidence: 98.2%
Identity: 28%
Fold: RPB6/omega subunit-like
Superfamily: RPB6/omega subunit-like
Family: RNA polymerase omega subunit
Phyre2
| 3 |
|
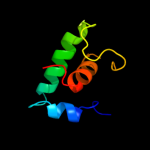
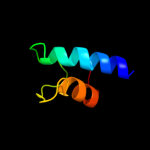
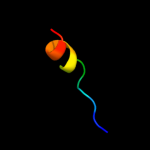
PDB 1smy chain E
Region: 2 - 60
Aligned: 59
Modelled: 59
Confidence: 98.1%
Identity: 25%
Fold: RPB6/omega subunit-like
Superfamily: RPB6/omega subunit-like
Family: RNA polymerase omega subunit
Phyre2
| 4 |
|
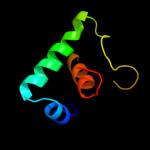
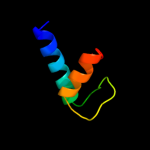
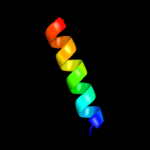
PDB 1twc chain F
Region: 15 - 60
Aligned: 45
Modelled: 46
Confidence: 97.0%
Identity: 22%
PDB header:transcription
Chain: F: PDB Molecule:dna-directed rna polymerases i, ii, and iii 23
PDBTitle: rna polymerase ii complexed with gtp
Phyre2
| 5 |
|
PDB 1twf chain F
Region: 15 - 60
Aligned: 45
Modelled: 46
Confidence: 97.0%
Identity: 22%
Fold: RPB6/omega subunit-like
Superfamily: RPB6/omega subunit-like
Family: RPB6
Phyre2
| 6 |
|
PDB 1qkl chain A
Region: 15 - 60
Aligned: 45
Modelled: 46
Confidence: 96.9%
Identity: 22%
Fold: RPB6/omega subunit-like
Superfamily: RPB6/omega subunit-like
Family: RPB6
Phyre2
| 7 |
|
PDB 3h0g chain F
Region: 15 - 60
Aligned: 45
Modelled: 46
Confidence: 96.9%
Identity: 22%
PDB header:transcription
Chain: F: PDB Molecule:dna-directed rna polymerases i, ii, and iii
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
Phyre2
| 8 |
|
PDB 2pmz chain W
Region: 15 - 60
Aligned: 45
Modelled: 46
Confidence: 96.8%
Identity: 27%
PDB header:translation, transferase
Chain: W: PDB Molecule:dna-directed rna polymerase subunit k;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
Phyre2
| 9 |
|
PDB 1hul chain A
Region: 18 - 57
Aligned: 38
Modelled: 40
Confidence: 17.7%
Identity: 21%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Short-chain cytokines
Phyre2
| 10 |
|
PDB 2kxh chain B
Region: 23 - 30
Aligned: 8
Modelled: 8
Confidence: 14.8%
Identity: 63%
PDB header:protein binding
Chain: B: PDB Molecule:peptide of far upstream element-binding protein 1;
PDBTitle: solution structure of the first two rrm domains of fir in the complex2 with fbp nbox peptide
Phyre2
| 11 |
|
PDB 1id3 chain A
Region: 41 - 54
Aligned: 14
Modelled: 14
Confidence: 13.5%
Identity: 43%
Fold: Histone-fold
Superfamily: Histone-fold
Family: Nucleosome core histones
Phyre2
| 12 |
|
PDB 1ysh chain E
Region: 14 - 31
Aligned: 18
Modelled: 18
Confidence: 11.8%
Identity: 33%
PDB header:structural protein/rna
Chain: E: PDB Molecule:40s ribosomal protein s13;
PDBTitle: localization and dynamic behavior of ribosomal protein l30e
Phyre2
| 13 |
|
PDB 1tzy chain C
Region: 45 - 54
Aligned: 10
Modelled: 10
Confidence: 7.5%
Identity: 50%
Fold: Histone-fold
Superfamily: Histone-fold
Family: Nucleosome core histones
Phyre2
| 14 |
|
PDB 1p3i chain E
Region: 41 - 54
Aligned: 14
Modelled: 14
Confidence: 6.6%
Identity: 36%
Fold: Histone-fold
Superfamily: Histone-fold
Family: Nucleosome core histones
Phyre2
| 15 |
|
PDB 3ci9 chain B
Region: 6 - 18
Aligned: 13
Modelled: 13
Confidence: 6.5%
Identity: 23%
PDB header:transcription
Chain: B: PDB Molecule:heat shock factor-binding protein 1;
PDBTitle: crystal structure of the human hsbp1
Phyre2