| 1 |
|
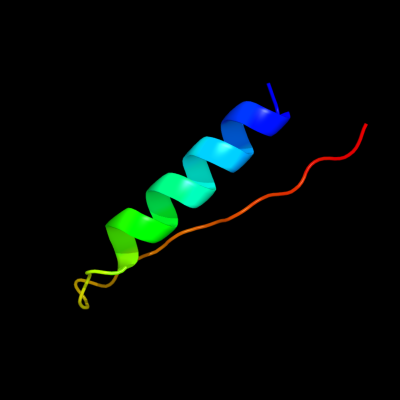
PDB 2rkh chain A
Region: 59 - 91
Aligned: 33
Modelled: 33
Confidence: 14.0%
Identity: 30%
PDB header:transcription
Chain: A: PDB Molecule:putative apha-like transcription factor;
PDBTitle: crystal structure of a putative apha-like transcription factor2 (zp_00208345.1) from magnetospirillum magnetotacticum ms-1 at 2.00 a3 resolution
Phyre2
| 2 |
|
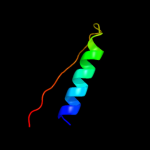
PDB 3bj6 chain B
Region: 4 - 37
Aligned: 34
Modelled: 34
Confidence: 13.3%
Identity: 12%
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcription regulator sp03579
Phyre2
| 3 |
|
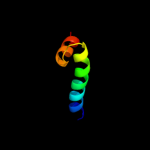
PDB 2voy chain G
Region: 77 - 102
Aligned: 26
Modelled: 26
Confidence: 11.1%
Identity: 15%
PDB header:hydrolase
Chain: G: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 4 |
|
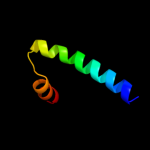
PDB 1gdh chain A domain 2
Region: 64 - 98
Aligned: 34
Modelled: 35
Confidence: 10.5%
Identity: 15%
Fold: Flavodoxin-like
Superfamily: Formate/glycerate dehydrogenase catalytic domain-like
Family: Formate/glycerate dehydrogenases, substrate-binding domain
Phyre2
| 5 |
|
PDB 2a61 chain A domain 1
Region: 2 - 37
Aligned: 36
Modelled: 36
Confidence: 8.7%
Identity: 22%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: MarR-like transcriptional regulators
Phyre2
| 6 |
|
PDB 1yhu chain A
Region: 7 - 72
Aligned: 65
Modelled: 66
Confidence: 8.0%
Identity: 14%
PDB header:oxygen storage/transport
Chain: A: PDB Molecule:hemoglobin a1 chain;
PDBTitle: crystal structure of riftia pachyptila c1 hemoglobin reveals novel2 assembly of 24 subunits.
Phyre2
| 7 |
|
PDB 2d2m chain B
Region: 7 - 69
Aligned: 63
Modelled: 63
Confidence: 7.5%
Identity: 16%
PDB header:oxygen storage/transport
Chain: B: PDB Molecule:giant hemoglobin, a2(a5) globin chain;
PDBTitle: structure of an extracellular giant hemoglobin of the2 gutless beard worm oligobrachia mashikoi
Phyre2
| 8 |
|
PDB 1t6f chain A
Region: 68 - 75
Aligned: 8
Modelled: 8
Confidence: 6.7%
Identity: 38%
PDB header:cell cycle
Chain: A: PDB Molecule:geminin;
PDBTitle: crystal structure of the coiled-coil dimerization motif of2 geminin
Phyre2
| 9 |
|
PDB 1q1f chain A
Region: 10 - 69
Aligned: 60
Modelled: 60
Confidence: 6.4%
Identity: 17%
Fold: Globin-like
Superfamily: Globin-like
Family: Globins
Phyre2