| 1 |
|
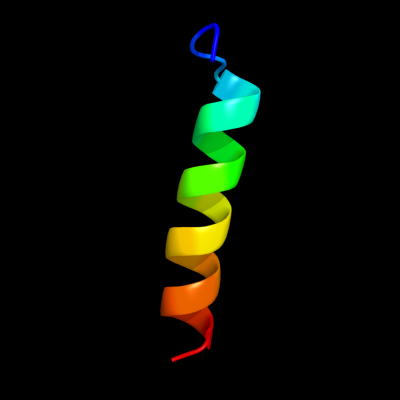
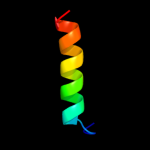
PDB 2juz chain A domain 1
Region: 57 - 78
Aligned: 22
Modelled: 22
Confidence: 22.5%
Identity: 14%
Fold: YejL-like
Superfamily: YejL-like
Family: YejL-like
Phyre2
| 2 |
|
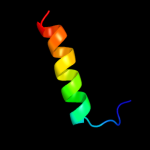
PDB 2qgp chain A
Region: 6 - 35
Aligned: 30
Modelled: 30
Confidence: 20.4%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:hnh endonuclease;
PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr87.
Phyre2
| 3 |
|
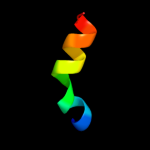
PDB 2rhf chain A
Region: 57 - 80
Aligned: 24
Modelled: 24
Confidence: 20.0%
Identity: 25%
PDB header:hydrolase
Chain: A: PDB Molecule:dna helicase recq;
PDBTitle: d. radiodurans recq hrdc domain 3
Phyre2
| 4 |
|
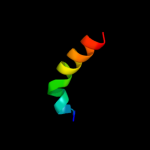
PDB 2rrd chain A
Region: 57 - 84
Aligned: 28
Modelled: 28
Confidence: 18.2%
Identity: 21%
PDB header:dna binding protein
Chain: A: PDB Molecule:hrdc domain from bloom syndrome protein;
PDBTitle: structure of hrdc domain from human bloom syndrome protein, blm
Phyre2
| 5 |
|
PDB 1rqp chain A domain 1
Region: 6 - 13
Aligned: 8
Modelled: 8
Confidence: 17.8%
Identity: 50%
Fold: Bacterial fluorinating enzyme, C-terminal domain
Superfamily: Bacterial fluorinating enzyme, C-terminal domain
Family: Bacterial fluorinating enzyme, C-terminal domain
Phyre2
| 6 |
|
PDB 2juw chain A domain 1
Region: 56 - 78
Aligned: 23
Modelled: 23
Confidence: 15.7%
Identity: 22%
Fold: YejL-like
Superfamily: YejL-like
Family: YejL-like
Phyre2
| 7 |
|
PDB 2jrx chain A domain 1
Region: 57 - 78
Aligned: 22
Modelled: 22
Confidence: 15.3%
Identity: 18%
Fold: YejL-like
Superfamily: YejL-like
Family: YejL-like
Phyre2
| 8 |
|
PDB 2jpq chain A domain 1
Region: 56 - 78
Aligned: 23
Modelled: 23
Confidence: 15.1%
Identity: 13%
Fold: YejL-like
Superfamily: YejL-like
Family: YejL-like
Phyre2
| 9 |
|
PDB 3khz chain A
Region: 61 - 77
Aligned: 17
Modelled: 17
Confidence: 7.5%
Identity: 18%
PDB header:hydrolase
Chain: A: PDB Molecule:putative dipeptidase sacol1801;
PDBTitle: crystal structure of r350a mutant of staphylococcus aureus2 metallopeptidase (sapep/dape) in the apo-form
Phyre2
| 10 |
|
PDB 1wud chain A domain 1
Region: 57 - 80
Aligned: 24
Modelled: 24
Confidence: 7.4%
Identity: 25%
Fold: SAM domain-like
Superfamily: HRDC-like
Family: HRDC domain from helicases
Phyre2