| 1 |
|
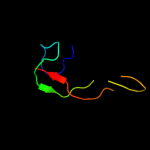
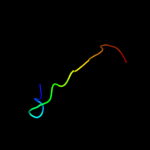
PDB 1k0i chain A domain 2
Region: 5 - 26
Aligned: 22
Modelled: 22
Confidence: 24.5%
Identity: 36%
Fold: FAD-linked reductases, C-terminal domain
Superfamily: FAD-linked reductases, C-terminal domain
Family: PHBH-like
Phyre2
| 2 |
|
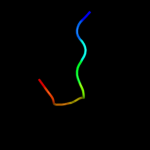
PDB 3muw chain U
Region: 11 - 53
Aligned: 40
Modelled: 41
Confidence: 15.7%
Identity: 30%
PDB header:virus
Chain: U: PDB Molecule:structural polyprotein;
PDBTitle: pseudo-atomic structure of the e2-e1 protein shell in sindbis virus
Phyre2
| 3 |
|
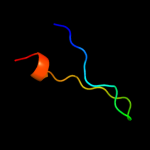
PDB 2jqo chain A
Region: 6 - 21
Aligned: 16
Modelled: 16
Confidence: 13.0%
Identity: 38%
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical protein yoba;
PDBTitle: nmr solution structure of bacillus subtilis yoba 21-120:2 northeast structural genomics consortium target sr547
Phyre2
| 4 |
|
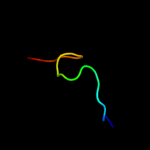
PDB 2ldj chain A
Region: 2 - 16
Aligned: 15
Modelled: 15
Confidence: 11.7%
Identity: 53%
PDB header:de novo protein
Chain: A: PDB Molecule:trp-cage mini-protein;
PDBTitle: 1h chemical shift assignments and structure of trp-cage mini-protein2 with d-amino acid
Phyre2
| 5 |
|
PDB 1cza chain N domain 3
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 9.7%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2
| 6 |
|
PDB 2ovf chain A
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 8.2%
Identity: 45%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:stal;
PDBTitle: crystal structure of stal-pap complex
Phyre2
| 7 |
|
PDB 1bg3 chain A domain 1
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 7.9%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2
| 8 |
|
PDB 1v4s chain A domain 1
Region: 4 - 19
Aligned: 16
Modelled: 16
Confidence: 7.8%
Identity: 38%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2
| 9 |
|
PDB 1bg3 chain A domain 3
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 7.0%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2
| 10 |
|
PDB 1osy chain A
Region: 15 - 43
Aligned: 29
Modelled: 29
Confidence: 6.5%
Identity: 31%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Fungal immunomodulatory protein, FIP
Family: Fungal immunomodulatory protein, FIP
Phyre2
| 11 |
|
PDB 2j44 chain A domain 1
Region: 20 - 38
Aligned: 19
Modelled: 19
Confidence: 6.4%
Identity: 42%
Fold: Prealbumin-like
Superfamily: Starch-binding domain-like
Family: PUD-like
Phyre2
| 12 |
|
PDB 2h8k chain A
Region: 7 - 15
Aligned: 9
Modelled: 9
Confidence: 6.3%
Identity: 78%
PDB header:transferase
Chain: A: PDB Molecule:sult1c3 splice variant d;
PDBTitle: human sulfotranferase sult1c3 in complex with pap
Phyre2
| 13 |
|
PDB 1ig8 chain A domain 1
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 6.3%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2
| 14 |
|
PDB 2bh7 chain A
Region: 25 - 33
Aligned: 9
Modelled: 9
Confidence: 6.1%
Identity: 56%
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
Phyre2
| 15 |
|
PDB 1q44 chain A
Region: 7 - 15
Aligned: 9
Modelled: 9
Confidence: 5.8%
Identity: 56%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: PAPS sulfotransferase
Phyre2
| 16 |
|
PDB 3f3h chain A
Region: 15 - 37
Aligned: 23
Modelled: 23
Confidence: 5.6%
Identity: 26%
PDB header:antitumor protein
Chain: A: PDB Molecule:immunomodulatory protein ling zhi-8;
PDBTitle: crystal structure and anti-tumor activity of lz-8 from the fungus2 ganoderma lucidium
Phyre2
| 17 |
|
PDB 1v58 chain A domain 2
Region: 5 - 36
Aligned: 32
Modelled: 32
Confidence: 5.3%
Identity: 28%
Fold: Cystatin-like
Superfamily: DsbC/DsbG N-terminal domain-like
Family: DsbC/DsbG N-terminal domain-like
Phyre2
| 18 |
|
PDB 1xcr chain A domain 1
Region: 27 - 43
Aligned: 13
Modelled: 17
Confidence: 5.3%
Identity: 77%
Fold: AF0104/ALDC/Ptd012-like
Superfamily: AF0104/ALDC/Ptd012-like
Family: PTD012-like
Phyre2
| 19 |
|
PDB 1cza chain N domain 1
Region: 5 - 19
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 40%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Hexokinase
Phyre2