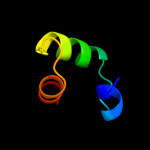
1 c1tr8A_
64.9
39
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
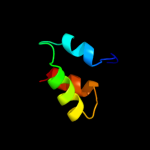
2 d1vega_
40.9
25
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain3 d1veja1
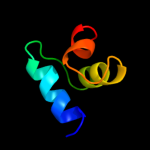
36.6
31
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain4 d2bwba1
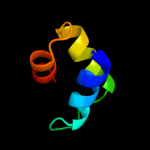
36.6
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain5 c2cwbA_
36.2
34
PDB header: protein bindingChain: A: PDB Molecule: chimera of immunoglobulin g binding protein gPDBTitle: solution structure of the ubiquitin-associated domain of2 human bmsc-ubp and its complex with ubiquitin
6 c2dahA_
35.8
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ubiquilin-3;PDBTitle: solution structure of the c-terminal uba domain in the2 human ubiquilin 3
7 d1vdla_
35.8
29
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain8 c1wr1B_
31.1
25
PDB header: signaling proteinChain: B: PDB Molecule: ubiquitin-like protein dsk2;PDBTitle: the complex sturcture of dsk2p uba with ubiquitin
9 c2jy5A_
30.2
31
PDB header: signaling proteinChain: A: PDB Molecule: ubiquilin-1;PDBTitle: nmr structure of ubiquilin 1 uba domain
10 d1ifya_
28.3
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain11 d1aipc1
28.1
47
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain12 c2dnaA_
27.6
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: unnamed protein product;PDBTitle: solution structure of rsgi ruh-056, a uba domain from mouse2 cdna
13 c2d9sA_
27.4
29
PDB header: ligaseChain: A: PDB Molecule: cbl e3 ubiquitin protein ligase;PDBTitle: solution structure of rsgi ruh-049, a uba domain from mouse2 cdna
14 d1oqya1
27.3
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain15 d2dnaa1
25.0
25
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain16 c2dchX_
25.0
50
PDB header: hydrolaseChain: X: PDB Molecule: putative homing endonuclease;PDBTitle: crystal structure of archaeal intron-encoded homing endonuclease i-2 tsp061i
17 d1xb2b1
24.1
35
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain18 d1z96a1
24.0
38
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain19 d2cp9a1
23.0
35
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain20 d1ir1s_
22.8
26
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit21 d2daha1
not modelled
21.9
28
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain22 d1efub3
not modelled
21.1
47
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain23 c2do6A_
not modelled
20.7
26
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of rsgi ruh-065, a uba domain from human2 cdna
24 d1rbli_
not modelled
18.6
37
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit25 c1umqA_
not modelled
16.5
17
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
26 d1umqa_
not modelled
16.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like27 d1etxa_
not modelled
15.8
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like28 d1uzhc1
not modelled
15.4
37
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit29 d1fipa_
not modelled
15.1
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like30 c3lwfD_
not modelled
15.1
15
PDB header: transcription regulatorChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator (np_470886.1)2 from listeria innocua at 2.06 a resolution
31 d1ngka_
not modelled
14.5
18
Fold: Globin-likeSuperfamily: Globin-likeFamily: Truncated hemoglobin32 c3d19E_
not modelled
14.1
25
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: conserved metalloprotein;PDBTitle: crystal structure of a conserved metalloprotein from bacillus cereus
33 d1bwvs_
not modelled
14.0
32
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit34 d1bxni_
not modelled
13.6
42
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit35 d1svdm1
not modelled
13.1
26
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit36 d1ylfa1
not modelled
12.9
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator Rrf237 c2jnhA_
not modelled
12.9
28
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of the uba domain from cbl-b
38 d1x38a2
not modelled
12.6
31
Fold: Flavodoxin-likeSuperfamily: Beta-D-glucan exohydrolase, C-terminal domainFamily: Beta-D-glucan exohydrolase, C-terminal domain39 d1wiva_
not modelled
12.1
33
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain40 c3zv0A_
not modelled
12.0
24
PDB header: cell cycleChain: A: PDB Molecule: protein shq1;PDBTitle: structure of the shq1p-cbf5p complex
41 d1etob_
not modelled
11.4
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like42 c3iv1F_
not modelled
11.0
47
PDB header: hydrolaseChain: F: PDB Molecule: tumor susceptibility gene 101 protein;PDBTitle: coiled-coil domain of tumor susceptibility gene 101
43 d1ejxc2
not modelled
10.6
34
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: alpha-subunit of urease, catalytic domain44 c2y75F_
not modelled
10.1
24
PDB header: transcriptionChain: F: PDB Molecule: hth-type transcriptional regulator cymr;PDBTitle: the structure of cymr (yrzc) the global cysteine regulator2 of b. subtilis
45 c2cosA_
not modelled
10.0
41
PDB header: transferaseChain: A: PDB Molecule: serine/threonine protein kinase lats2;PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
46 d1id3c_
not modelled
10.0
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones47 c3ju8B_
not modelled
9.7
10
PDB header: oxidoreductaseChain: B: PDB Molecule: succinylglutamic semialdehyde dehydrogenase;PDBTitle: crystal structure of succinylglutamic semialdehyde dehydrogenase from2 pseudomonas aeruginosa.
48 c2bmmA_
not modelled
9.5
15
PDB header: oxygen storage/transportChain: A: PDB Molecule: thermostable hemoglobin from thermobifida fusca;PDBTitle: x-ray structure of a novel thermostable hemoglobin from the2 actinobacterium thermobifida fusca
49 d1v92a_
not modelled
9.4
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TAP-C domain-like50 d1bdfa1
not modelled
9.4
24
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain51 d8ruci_
not modelled
9.1
26
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit52 d2ezia_
not modelled
8.9
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain53 d1k3ra1
not modelled
8.4
42
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Hypothetical protein MTH1 (MT0001), insert domain54 c2dakA_
not modelled
8.4
24
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin carboxyl-terminal hydrolase 5;PDBTitle: solution structure of the second uba domain in the human2 ubiquitin specific protease 5 (isopeptidase 5)
55 d1wdds_
not modelled
8.1
26
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit56 d1e9yb2
not modelled
8.1
28
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: alpha-subunit of urease, catalytic domain57 d1q8ba_
not modelled
8.1
27
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Hypothetical protein YjcS58 d2ezha_
not modelled
8.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain59 d1gk8i_
not modelled
8.0
37
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit60 c1zvaA_
not modelled
8.0
64
PDB header: viral proteinChain: A: PDB Molecule: e2 glycoprotein;PDBTitle: a structure-based mechanism of sars virus membrane fusion
61 d2v6ai1
not modelled
7.9
37
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit62 d2f2ga1
not modelled
7.8
43
Fold: Heme oxygenase-likeSuperfamily: Heme oxygenase-likeFamily: TENA/THI-463 c3f44A_
not modelled
7.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: putative monooxygenase;PDBTitle: crystal structure of putative monooxygenase (yp_193413.1) from2 lactobacillus acidophilus ncfm at 1.55 a resolution
64 c3iukB_
not modelled
7.2
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of putative bacterial protein of unknown function2 (duf885, pf05960.1, ) from arthrobacter aurescens tc1, reveals fold3 similar to that of m32 carboxypeptidases
65 d1ej7s_
not modelled
7.1
26
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit66 d2jssa1
not modelled
7.0
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones67 d1uzdc1
not modelled
7.0
37
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit68 c1ygmA_
not modelled
7.0
47
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein bsu31320;PDBTitle: nmr structure of mistic
69 d2hkja2
not modelled
6.9
36
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain70 d2f5va2
not modelled
6.9
20
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: GMC oxidoreductases71 c2f8nK_
not modelled
6.8
33
PDB header: structural protein/dnaChain: K: PDB Molecule: histone h2a type 1;PDBTitle: 2.9 angstrom x-ray structure of hybrid macroh2a nucleosomes
72 c3lk6A_
not modelled
6.8
20
PDB header: hydrolaseChain: A: PDB Molecule: lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
73 d1ntca_
not modelled
6.7
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like74 c3e7lD_
not modelled
6.6
10
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
75 d1tzya_
not modelled
6.6
33
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones76 c2kzhA_
not modelled
6.5
13
PDB header: isomeraseChain: A: PDB Molecule: tryptophan biosynthesis protein trpcf;PDBTitle: three-dimensional structure of a truncated phosphoribosylanthranilate2 isomerase (residues 255-384) from escherichia coli
77 d1kx3c_
not modelled
6.5
33
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones78 d1u35c1
not modelled
6.4
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones79 c3cxjB_
not modelled
6.4
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from2 methanothermobacter thermautotrophicus
80 d2g3qa1
not modelled
6.3
34
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain81 c2rdxG_
not modelled
6.2
21
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: mandelate racemase/muconate lactonizing enzyme, putative;PDBTitle: crystal structure of mandelate racemase/muconate lactonizing enzyme2 from roseovarius nubinhibens ism
82 c2o4wA_
not modelled
6.1
31
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme;PDBTitle: t4 lysozyme circular permutant
83 c3exqA_
not modelled
6.1
26
PDB header: hydrolaseChain: A: PDB Molecule: nudix family hydrolase;PDBTitle: crystal structure of a nudix family hydrolase from2 lactobacillus brevis
84 d1f66c_
not modelled
6.0
27
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones85 c1f66C_
not modelled
6.0
27
PDB header: structural protein/dnaChain: C: PDB Molecule: histone h2a.z;PDBTitle: 2.6 a crystal structure of a nucleosome core particle2 containing the variant histone h2a.z
86 d2cosa1
not modelled
5.9
41
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain87 d1cuka1
not modelled
5.9
50
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain88 c1xb2B_
not modelled
5.8
35
PDB header: translationChain: B: PDB Molecule: elongation factor ts, mitochondrial;PDBTitle: crystal structure of bos taurus mitochondrial elongation2 factor tu/ts complex
89 c2jssA_
not modelled
5.8
27
PDB header: chaperone/nuclear proteinChain: A: PDB Molecule: chimera of histone h2b.1 and histone h2a.z;PDBTitle: nmr structure of chaperone chz1 complexed with histone2 h2a.z-h2b
90 c3osyA_
not modelled
5.8
41
PDB header: hydrolaseChain: A: PDB Molecule: 3c protease;PDBTitle: human enterovirus 71 3c protease
91 d1hioa_
not modelled
5.7
33
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones92 d1mxaa1
not modelled
5.7
23
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase93 d1eqza_
not modelled
5.6
33
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones94 d2p02a1
not modelled
5.5
31
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase95 d1qm4a1
not modelled
5.5
23
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase96 d1euha_
not modelled
5.5
17
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like97 c3kn1A_
not modelled
5.4
21
PDB header: protein bindingChain: A: PDB Molecule: golgi phosphoprotein 3;PDBTitle: crystal structure of golgi phosphoprotein 3 n-term2 truncation variant
98 c3c38A_
not modelled
5.4
11
PDB header: signaling protein, transferaseChain: A: PDB Molecule: autoinducer 2 sensor kinase/phosphatase luxq;PDBTitle: crystal structure of the periplasmic domain of vibrio2 cholerae luxq
99 c3bmxB_
not modelled
5.4
20
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis