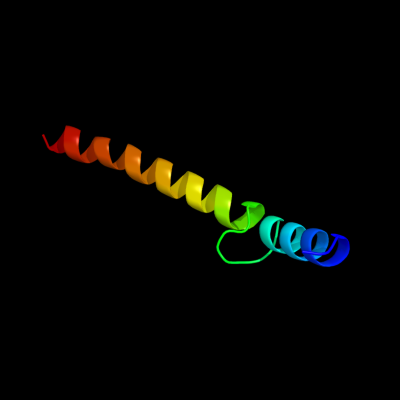
1 c3sokB_
95.6
24
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
2 d1oqwa_
94.5
8
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin3 d2pila_
91.6
17
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin4 c2ky8A_
62.2
17
PDB header: transcription/dnaChain: A: PDB Molecule: methyl-cpg-binding domain protein 2;PDBTitle: solution structure and dynamic analysis of chicken mbd2 methyl binding2 domain bound to a target methylated dna sequence
5 d1ig4a_
55.0
12
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD6 d1qk9a_
44.7
21
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD7 d1ub1a_
34.0
21
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD8 d1w8oa1
28.3
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes9 d3ci0i1
25.6
7
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: GSPII I/J protein-like10 d2reta1
18.9
10
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: GSPII I/J protein-like11 d1yfua1
15.7
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like12 c2jv4A_
13.4
20
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis/trans isomerase;PDBTitle: structure characterisation of pina ww domain and comparison2 with other group iv ww domains, pin1 and ess1
13 d2uuyb1
11.4
27
Fold: BPTI-likeSuperfamily: BPTI-likeFamily: Tick tryptase inhibitor-like14 c1ymzA_
10.2
13
PDB header: unknown functionChain: A: PDB Molecule: cc45;PDBTitle: cc45, an artificial ww domain designed using statistical2 coupling analysis
15 c3nctC_
9.7
20
PDB header: dna binding protein, chaperoneChain: C: PDB Molecule: protein psib;PDBTitle: x-ray crystal structure of the bacterial conjugation factor psib, a2 negative regulator of reca
16 c1e0mA_
9.7
14
PDB header: de novo proteinChain: A: PDB Molecule: wwprototype;PDBTitle: prototype ww domain
17 c1wr4A_
9.7
13
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-protein ligase nedd4-2;PDBTitle: solution structure of the second ww domain of nedd4-2
18 c1dvaY_
9.3
50
PDB header: hydrolase/hydrolase inhibitorChain: Y: PDB Molecule: peptide e-76;PDBTitle: crystal structure of the complex between the peptide exosite inhibitor2 e-76 and coagulation factor viia
19 c1dvaX_
9.3
50
PDB header: hydrolase/hydrolase inhibitorChain: X: PDB Molecule: peptide e-76;PDBTitle: crystal structure of the complex between the peptide exosite inhibitor2 e-76 and coagulation factor viia
20 c2retE_
8.9
15
PDB header: protein transportChain: E: PDB Molecule: pseudopilin epsi;PDBTitle: the crystal structure of a binary complex of two pseudopilins: epsi2 and epsj from the type 2 secretion system of vibrio vulnificus
21 c2ysbA_
not modelled
8.1
20
PDB header: protein bindingChain: A: PDB Molecule: salvador homolog 1 protein;PDBTitle: solution structure of the first ww domain from the mouse2 salvador homolog 1 protein (sav1)
22 d1tk7a2
not modelled
7.6
13
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain23 d2jmfa1
not modelled
7.5
15
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain24 d1is3a_
not modelled
7.5
8
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)25 c1yiuA_
not modelled
7.3
13
PDB header: ligaseChain: A: PDB Molecule: itchy e3 ubiquitin protein ligase;PDBTitle: itch e3 ubiquitin ligase ww3 domain
26 d1k9ra_
not modelled
7.1
20
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain27 d2e9ia1
not modelled
7.0
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)28 d2cs3a1
not modelled
6.8
20
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: Variant RING domain29 c2yshA_
not modelled
6.5
18
PDB header: protein bindingChain: A: PDB Molecule: growth-arrest-specific protein 7;PDBTitle: solution structure of the ww domain from the human growth-2 arrest-specific protein 7, gas-7
30 c2y69Z_
not modelled
6.3
8
PDB header: electron transportChain: Z: PDB Molecule: cytochrome c oxidase polypeptide 8h;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
31 c3a0hl_
not modelled
6.3
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
32 c3a0hL_
not modelled
6.3
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
33 c1wr7A_
not modelled
6.3
13
PDB header: ligaseChain: A: PDB Molecule: nedd4-2;PDBTitle: solution structure of the third ww domain of nedd4-2
34 c3a0bl_
not modelled
6.2
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
35 c3a0bL_
not modelled
6.2
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
36 c2lawA_
not modelled
6.0
23
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: yorkie homolog;PDBTitle: structure of the second ww domain from human yap in complex with a2 human smad1 derived peptide
37 d2axtl1
not modelled
6.0
30
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like38 c1s5lL_
not modelled
6.0
30
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
39 c1s5ll_
not modelled
6.0
30
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
40 c2axtL_
not modelled
6.0
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
41 c2axtl_
not modelled
6.0
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
42 c3bz1L_
not modelled
6.0
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
43 c3arcL_
not modelled
6.0
30
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
44 c3prqL_
not modelled
6.0
30
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
45 c3kziL_
not modelled
6.0
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
46 c3prrL_
not modelled
6.0
30
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
47 c3bz2L_
not modelled
6.0
30
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
48 d2ysca1
not modelled
6.0
15
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain49 c3u5eL_
not modelled
5.9
35
PDB header: ribosomeChain: L: PDB Molecule: 60s ribosomal protein l13-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
50 c3arcl_
not modelled
5.6
30
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
51 c2dmvA_
not modelled
5.6
18
PDB header: ligaseChain: A: PDB Molecule: itchy homolog e3 ubiquitin protein ligase;PDBTitle: solution structure of the second ww domain of itchy homolog2 e3 ubiquitin protein ligase (itch)
52 c2ez5W_
not modelled
5.5
13
PDB header: signalling protein,ligaseChain: W: PDB Molecule: e3 ubiquitin-protein ligase nedd4;PDBTitle: solution structure of the dnedd4 ww3* domain- comm lpsy2 peptide complex
53 c2djyA_
not modelled
5.4
20
PDB header: ligase/signaling proteinChain: A: PDB Molecule: smad ubiquitination regulatory factor 2;PDBTitle: solution structure of smurf2 ww3 domain-smad7 py peptide2 complex
54 c2wsfG_
not modelled
5.4
25
PDB header: photosynthesisChain: G: PDB Molecule: photosystem i reaction center subunit v,PDBTitle: improved model of plant photosystem i
55 c2yscA_
not modelled
5.3
15
PDB header: protein bindingChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding familyPDBTitle: solution structure of the ww domain from the human amyloid2 beta a4 precursor protein-binding family b member 3, apbb3